Abstract
Designing anticoronavirus disease 2019 (anti-COVID-19) agents is the primary concern of medicinal chemists/drug designers nowadays. Repurposing of known active compounds against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new effective and time-saving trend in anti-COVID-19 drug discovery. Thorough inhibition of the coronaviral-2 proteins (i.e., multitarget inhibition) is a possible powerful favorable strategy for developing effectively potent drugs for COVID-19. In this new research study, I succeeded to repurpose the two antioxidant polyhydroxy-1,3,4-oxadiazole compounds CoViTris2020 and ChloViD2020 as the first multitarget coronaviral protein blockers with extremely higher potencies (reach about 65 and 304 times, for CoViTris2020, and 20 and 93 times, for ChloViD2020, more potent than remdesivir and favipiravir, respectively). These two 2,5-disubstituted-1,3,4-oxadiazoles were computationally studied (through molecular docking in almost all SARS-CoV-2 proteins) and biologically assessed (through a newly established robust in vitro anti-COVID-19 assay) for their anticoronaviral-2 bioactivities. The data obtained from the docking investigation showed that both ligands promisingly exhibited very strong inhibitory binding affinities with almost all docked enzymes (e.g., they displayed extremely lower binding energies of − 12.00 and − 9.60 kcal/mol, respectively, with the SARS-CoV-2 RNA-dependent RNA polymerase “RdRp”). The results of the biological assay revealed that CoViTris2020 and ChloViD2020 significantly displayed very high anti-COVID-19 activities (anti-SARS-CoV-2 EC50 = 0.31 and 1.01 μM, respectively). Further in vivo/clinical studies for the development of CoViTris2020 and ChloViD2020 as anti-COVID-19 medications are required. In brief, the ascent of CoViTris2020 and ChloViD2020 as the two lead members of the novel family of anti-COVID-19 polyphenolic 2,5-disubstituted-1,3,4-oxadiazole derivatives represents a promising hope in COVID-19 therapy.
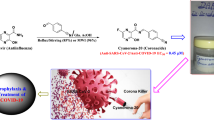
Graphic abstract
CoViTris2020 and ChloViD2020 inhibit SARS-CoV-2 life cycle with surprising EC50 values of 0.31 and 1.01 μM, respectively. CoViTris2020 strongly inhibits coronaviral-2 RdRp with exceptionally lower inhibitory binding energy of − 12.00 kcal/mol.

Similar content being viewed by others
Introduction
A broad variety of tactics and strategies was employed to fight the current worldwide coronavirus disease 2019 (COVID-19) epidemic as an urgent result of the novelty of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections, and the lack of effective medications [1,2,3]. Many plans of these emerging strategies depend on reevaluating and repurposing existent drugs, and some others are totally new [4]. The design in each case depends on present scientific proof and evidence of logic mechanistic approaches that are efficient against either analogous/identical viral infections or the severe signs/symptoms that are caused by COVID-19 [1,2,3,4]. Comprehensive inhibition of the coronaviral-2 proteins (i.e., multitarget inhibition) can establish one of the most promising plans for developing effectively potent drugs for COVID-19. However, all the discovered blockers of the main stages of SARS-CoV-2 life cycle lack the required maximal potencies against the major SARS-CoV-2 enzymes (e.g., RNA-dependent RNA polymerase “RdRp”, papain-like protease “PLpro”, and main protease “Mpro”) [3, 4].
Many known drugs are currently under anti-COVID-19 therapeutic investigation, such as remdesivir (antiviral), ivermectin (antiparasitic), favipiravir (antiinfluenza), hydroxychloroquine (antimalarial and antirheumatic disorders), and arbidol (antiinfluenza) [4,5,6,7,8,9,10,11,12,13,14,15,16,17]. Perspectives of many medicinal chemists are based on utilizing the in silico computational molecular modeling studies as the starting points for designing novel potent compounds active against SARS-CoV-2, and most of these studies have been collected in currently published reviews [18, 19]. This current direction in searching for effective safe known compounds having the structural flexibility to be successfully redirected and repurposed as potential agents against COVID-19 triggered me to screen our library of known compounds/drugs (including my previously synthesized active compounds). Surprisingly, the predictive computational molecular search and screening gave rise to the discovery of two promising compounds in respect to the anti-SARS-CoV-2 activities, 1,2,3-tris[5-(3,4,5-trihydroxyphenyl)-1,3,4-oxadiazol-2-yl]propan-2-ol (CoViTris2020) and 5-[5-(7-chloro-4-hydroxyquinolin-3-yl)-1,3,4-oxadiazol-2-yl]benzene-1,2,3-triol (ChloViD2020) (Fig. 1). It is worth noticing that both compounds were previously synthesized and evaluated as effective antioxidant agents [20].
The adaptable and flexible scaffolds of CoViTris2020 and ChloViD2020 molecules are characterized by carrying significantly considerable number of hydrogen bond donors/acceptors and aromatic rings, which are the principal key moieties required for the best/ideal/strongest binding (hydrogen bonding and hydrophobic interactions) with the binding domains of almost all the COVID-19 protein targets in SARS-CoV-2 and human. It is worth mentioning that CoViTris2020 and ChloViD2020 are chemically very analogous to RNA nucleos(t)ides, giving them the expected abilities of being efficient bioisosteric antimetabolites in COVID-19 treatment. Additionally, CoViTris2020 and ChloViD2020 structures are also characterized by their abilities to strongly bound to zinc atoms, where they have many active zincophoric centers (i.e., they are rich in oxygenous and nitrogenous moieties) allowing them to act as strong multizincophores. Zinc atom/ion carriers have been extensively studied especially for their potent antiviral activities and they have been found to effectively inhibit the replication/reproduction machinery of several viruses in vitro [21]. Specifically, Zn2+ ions in certain exceedingly higher/lower concentrations can impair and disrupt the coronaviral replication and transcription and, therefore, excessive displacement or transport of Zn2+ ions by zincophoric agents (e.g., CoViTris2020 and ChloViD2020) will undoubtedly affect the coronaviral replication processes (zinc ionophores have been found to successfully and significantly inhibit the replication process of coronaviruses intracellularly in cell cultures) [10, 15, 22]. The considerable chemical resonance of both structures aids in boosting the comprehensive potential anti-COVID-19 bioactivities through intensifying the inhibitory chemical stabilities of the complexes of each compound with the active site residues of the coronaviral-2/human target proteins [20]. It is also worth noticing that the potent antioxidant actions of both compounds may aid their anti-SARS-CoV-2/anti-COVID-19 bioactivities through reducing almost all the oxidizing moieties of all the target proteins, which logically causes deactivation of the catalytic and noncatalytic biological activities of these important active proteins, meantime, these antioxidant properties aid in quenching the free radicals and fighting the oxidative stress inside the body of the COVID-19 patient [3, 20]. The existence of the bioactive antiviral 1,3,4-oxadiazole rings in the backbones of the two molecules is suggested to significantly enhance the anti-COVID-19 actions of both of them [20]. The two compounds have very balanced (ideal) lipophilic/hydrophilic properties (they have log P < 5, which perfectly obeys Lipinski’s Rule of Five “Ro5”) which are required to afford the top tolerated pharmacokinetic characteristics [23]. The two molecules were docked in all the available SARS-CoV-2 nonstructural and structural protein targets (including functional enzymes and proteins) along with one human structural enzyme, with an emphasis on the most important enzymes like the RdRp.
After deep investigative analysis of the SARS-CoV-2 constructure, researchers have been able to identify all the possible targets of the SARS-CoV-2 in a detailed way [24]. The promising results of the computational molecular docking of both compounds in almost all targets, in comparison with the reference potent drugs, theoretically confirmed their exceptional multitarget inhibitory activity against coronaviral-2 particles (to the best of my knowledge, the two compounds are considered the first potent multitarget anti-COVID-19 compounds). This interesting outcome inspired me to experimentally bioevaluate them. Precisely as the hypothetical predictions, the in vitro bioassay outcomes were also very promising. Based on all the previous facts and results, we can expect that CoViTris2020 and ChloViD2020 will successfully act as very potent anti-COVID-19 drug candidates through several and distinct mechanisms of action (i.e., through a very effective anti-COVID-19 mode of multiaction). Detailed illustrations of all the discovered target COVID-19 therapy nonstructural proteins (nsps) and structural proteins (sps) present in SARS-CoV-2 and human (up to date) are presented in Figs. 2 and 3, respectively. In this research paper, I report the successful repurposing of the two previously synthesized antioxidant 1,3,4-oxadiazole compounds, CoViTris2020 and ChloViD2020, as effective and potent anti-COVID-19 agents (as the first potent antidotal multitarget anti-SARS-CoV-2/anti-COVID-19 drugs).
Methods
Computational molecular docking studies of CoViTris2020 and ChloViD2020 (predictive anti-COVID-19 properties evaluation)
To expectedly assess the anti-COVID-19 activities of the two target antioxidant compounds of the current research, CoViTris2020 and ChloViD2020, prior to their actual practical anti-COVID-19 biological evaluation (preliminary in vitro/in vivo assays and subsequent preclinical/clinical trials), molecular docking of the molecules of both compounds in the target SARS-CoV-2 nsps and sps (along with one target human sp, ACE2) has been carried out using the docking engines of the most known and credible international molecular docking software programs (e.g., Discovery Studio, LeDock, GemDock, and GOLD). Different docking software programs were used to confidently and doubtlessly ensure the results and to assess and guarantee their reproducibility. Integrating the expected pharmacophoric features with the interaction energy analysis disclosed functionally pivotal amino acid residues in the binding pockets/cavities of the active and/or allosteric sites of the target SARS-CoV-2 enzymes (mainly) along with in silico predicted common inhibitory binding modes with the highly potent reference repurposed compounds (e.g., remdesivir, ivermectin, and favipiravir). The docking results were very brilliant to encourage me to utilize a new specific web-based server designed in 2020, which is specialized in precise molecular docking of all COVID-19 target nsps/sps and prediction of anti-COVID-19 activities and potencies of tested compounds. For this objective, the newly programmed COVID-19 Docking Server was used [25].
COVID-19 Docking Server web-based software (AutoDock Vina is used as the main docking engine) is an interactive web server for docking small molecules, peptides, or antibodies against potential protein targets of COVID-19 in order to predict the binding modes between COVID-19 targets and the ligands along with screening and evaluating the anti-COVID-19 activities of these ligands (i.e., the platform provides a free interactive modern tool using a very precise knowledge-based scoring function to evaluate the candidate binding poses for the specific prediction of COVID-19 target-ligand interactions and the following drug discovery for COVID-19). Generally, almost all SARS-CoV-2 nsps (enzymes) and very few SARS-CoV-2/human sps as COVID-19 therapy targets are recommended for small molecule docking. The structures of all the functional or structural protein targets involved in the SARS-CoV-2 replication life cycle were collected by direct downloading from international web databases (e.g., from the Protein Data Bank “PDB”) or constructed based on their known homologs of coronaviruses (by using homology modeling module of Maestro 10, website: www.schrodinger.com), and completely prepared ready for direct docking on this web-based software. The utmost effective and influenced nsps and sps (almost are functional enzymes) among all the detected and recognized proteins involved in COVID-19 therapy (previously presented in Figs. 2 and 3) were accurately opted to be targeted and docked. This includes fifteen various nCoV protein targets and one human protein target (the human enzyme ACE2). For docking of only one small molecule, the “Docking” mode box should be specifically selected as the computational module (type) for every specific target (this is the selection used in the current case). To get the best accurate results, an average exhaustiveness option of “12” was used. The detailed results of these estimations (binding energies in kcal/mol) are shown in Table 1. After docking, the structure of each enzyme/protein-compound complex was further examined and accurately analyzed for characterization by the help of the fully automated comprehensive interactive tool of the famous Protein–Ligand Interaction Profiler (PLIP) web server (https://projects.biotec.tu-dresden.de/plip-web) [26], and results were marginally tabulated for comparison, explication of the previous docking results, and then placing conclusions (see Table 2 as an example).
Anti-COVID-19 biological activities (in vitro assay) of CoViTris2020 and ChloViD2020
This novel and highly reliable anti-COVID-19 in vitro assay is based upon the authentic procedures of Chu and coworkers with very slight modifications [5, 27]. The complete procedures were carried out in a specialized biosafety level 3 (BSL-3) laboratory (SARS-CoV-2 is classified as a BSL-3 pathogen by the WHO and FDA) in Hong Kong SAR (China). The assayed SARS-CoV-2 virus, BetaCoV/Hong Kong/VM20001061/2020, was isolated from the fresh nasopharynx aspirate and throat swab of a confirmed fifty-years-aged COVID-19 male patient in Hong Kong using Vero E6 cells (ATCC CRL-1586). Stock virus (107.25 TCID50/mL) was prepared after three serial passages in Vero E6 cells in infection media (DMEM supplemented with 4.5 g/L D-glucose, 100 mg/L sodium pyruvate, 2% FBS, 100,000 U/L Penicillin–Streptomycin, and 25 mM HEPES). Following the original procedures in the literature, CoViTris2020 and ChloViD2020 compounds were synthesized (starting from gallic acid), purified (> 97% purity), fully characterized, and put in small dark brown glass containers to be ready for the assay [20]. The pure three reference compounds were purchased from MedChemExpress (remdesivir), Sigma-Aldrich (ivermectin, B1a form), and Toyama Chemical “Fujifilm group, Japan” (favipiravir). The stocks of the five tested compounds were accurately prepared by dissolving each of them in dimethylsulfoxide “DMSO” (to get a concentration of 100 mM of each of CoViTris2020, ChloViD2020, remdesivir, ivermectin, and favipiravir). To evaluate the in vitro anti-SARS-CoV-2 activities of the two target new compounds (CoViTris2020 and ChloViD2020) in comparison with those of the standard three reference compounds (mentioned above), Vero E6 cells were pretreated with the five compounds diluted in infection media for 1 h prior to infection by SARS-CoV-2 virus at MOI = 0.02. The five tested compounds were maintained with the virus inoculum during the 2-h incubation period. The inoculum was removed after incubation, and the cells were overlaid with infection media containing the diluted test compounds. After 48 h incubation at 37 °C, supernatants were immediately collected to quantify viral loads by TCID50 assay or quantitative real-time RT-PCR (TaqMan™ Fast Virus 1-Step Master Mix) [5, 27]. Viral loads in this assay were fitted in logarithm scale (log10 TCID50/mL and log10 viral RNA copies/mL), not in linear scale, under increasing concentrations of the tested compounds [5, 9, 27]. Four-parameter logistic regression (GraphPad Prism) was used to fit the dose–response curves and determine the EC50 of the tested compounds that inhibit SARS-CoV-2 viral replication (CPEIC100 was also determined for each compound). Cytotoxicity of each of the five tested compounds was evaluated in Vero E6 cells using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega) [5, 28]. The detailed values resulted from the previous assays (compound concentrations in μM) are shown in Table 3. Final results were represented as the mean ± SD from the triplicate biological experiments. Statistical analysis was performed using SkanIt 4.0 Research Edition software (ThermoFisher Scientific) and Prism V5 software (GraphPad). All reported data were significant at p < 0.05.
Results and discussion
Computational molecular docking studies of CoViTris2020 and ChloViD2020 (predictive anti-COVID-19 properties evaluation)
The deep understanding of the COVID-19 target-ligand interactions represents an extremely crucial key challenge in drug discovery for COVID-19. The computational simulative prediction of the SARS-CoV-2 proteins-inhibiting properties of the newly repurposed target compounds CoViTris2020 and ChloViD2020 greatly helped me to have a comprehensive overview of the expected anti-COVID-19 activities of the two compounds. This prediction also aided me to have a detailed conception about the two target compounds anti-COVID-19 modes of action along with the compounds degrees of efficacy and potency.
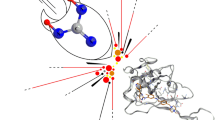
On close inspection of the top pose score values (present in Table 1) of docking most nsps and sps of SARS-CoV-2 (along with ACE2 of human, which is almost the only human entry point for the virus into the human body until now, see Introduction section and Fig. 3) using COVID-19 Docking Server, it is clearly observed that CoViTris2020 is distinguishably ranked first in its inhibitory binding affinities and potencies with an excellent range of binding energies of − 8.10 to − 12.00 kcal/mol. The binding affinities of CoViTris2020 dramatically exceed those of all the three reference drugs (they are about to be twice as those of favipiravir) and ChloViD2020, as the potent antioxidant CoViTris2020 molecule powerfully binds to the respective proteins forming very stable inhibited (deactivated) complexes with relatively amazing binding energies which are the lowest among all (i.e., significantly lower than the binding energies of all the other four compounds in their complexes with the respective nsps and sps). Specifically, CoViTris2020 molecule gives its best inhibitory binding affinities with the three nsps RdRp-RNA (RTP site) (− 12.00 kcal/mol), nsp14 (N7-MTase) (− 11.40 kcal/mol), and PLpro (dimer) (− 10.60 kcal/mol), respectively (Fig. 4a–d), which are three of the most effective protein targets in hindering and stopping the life cycle of the SARS-CoV-2 through inhibition and deactivation of their active sites [24]. These exceptional results are very promising as they indicate the high possibility of CoViTris2020 to be a very potent inhibitor (blocker) of RdRp, nsp14, and/or PLpro. Other proteins affording very encouraging excellent binding affinities with CoViTris2020 molecule include ACE2, nsp16 (all sites), N protein (NCB site), nsp3 (both sites), helicase (both sites), Mpro, and RdRp without RNA (RNA site) (Fig. 4b), respectively.
Screenshots of COVID-19 Docking Server outputs of the top predicted binding model of docking of CoViTris2020 molecule (colored pink) in: a SARS-CoV-2 RdRp-RNA “RTP site” (PDB code: 7BV2; colored with other various colors; Cartoon Style). b SARS-CoV-2 RdRp “RNA site” (PDB code: 7BV2; colored with other various colors; Trace Style). c SARS-CoV-2 nsp14 “N7-MTase” (PDB code: 5C8S, 1J53 “for active site homology”; colored with other various colors; Cartoon Style). d SARS-CoV-2 PLpro “dimer” (PDB code: 6WUU; colored with other various colors; Trace Style)
ChloViD2020 comes second, among all the five compounds, in its inhibitory binding energies and affinities which are ranged from − 7.50 to − 9.90 kcal/mol. The binding affinities of ChloViD2020 surpass those of all the three reference drugs (except for those of each of exoribonuclease and endoribonuclease with remdesivir, as there is binding energies difference of − 0.20 kcal/mol in favor of remdesivir in both cases), as the potent antioxidant ChloViD2020 molecule strongly binds to the respective proteins forming very stable inhibited complexes with relatively lower binding energies (i.e., significantly lower than the binding energies of all the three reference molecules in their complexes with the respective nsps and sps). ChloViD2020 molecule specifically gives its best inhibitory binding affinities with the four nsps nsp3 (207-379, MES site) (− 9.90 kcal/mol), RdRp-RNA (RTP site) (− 9.60 kcal/mol), nsp14 (N7-MTase) (− 9.30 kcal/mol), and PLpro (dimer) (− 9.30 kcal/mol), respectively (Fig. 5a–d). Interestingly, ChloViD2020 approaches and reaches the lower binding affinity of CoViTris2020 in only one enzyme which is N7-MTase (it is also one of the most effective protein targets in stopping the coronaviral-2 life cycle through blocking of its active site [24]) with the same binding energy value of -9.90 kcal/mol. These promising results indicate the significant possibility of ChloViD2020 to be a very potent inhibitor ligand or antagonist of nsp3, RdRp, nsp14, and/or PLpro. Other proteins giving very encouraging binding affinities with ChloViD2020 molecule include ACE2, N protein (NCB site), helicase (both sites), nsp16 (all sites), Mpro, Nsp15 (endoribonuclease), and RdRp without RNA (RNA site), respectively.
Screenshots of COVID-19 Docking Server outputs of the top predicted binding model of docking of ChloViD2020 molecule (colored pink) in: a SARS-CoV-2 nsp3 “207-379, MES site” (PDB code: 6W6Y; colored with other various colors; Cartoon Style). b SARS-CoV-2 RdRp-RNA “RTP site” (PDB code: 7BV2; colored with other various colors; Cartoon Style). c SARS-CoV-2 nsp14 “N7-MTase” (PDB code: 5C8S, 1J53 “for active site homology”; colored with other various colors; Cartoon Style). d SARS-CoV-2 PLpro “dimer” (PDB code: 6WUU; colored with other various colors; Trace Style)
The chemical structures of CoViTris2020 and ChloViD2020 molecules are exceptionally characterized by higher degrees of balanced orientational and conformational flexibility when compared to those of the other or reference anticoronaviral drugs (e.g., favipiravir, remdesivir, hydroxychloroquine, ivermectin, and arbidol). These extraordinary unique flexibilities of CoViTris2020 and ChloViD2020 structures are apparently observed in the docking poses in SARS-CoV-2 target proteins (along with human target ACE2) as shown in both Figs. 6 and 7, respectively. CoViTris2020 molecule (a trisubstituted or three armed bulky derivative of citric acid) has higher degree of flexibility than ChloViD2020 molecule (a typical 2,5-disubstituted derivative of 1,3,4-oxadiazole ring) due to, mainly, its larger topological polar surface area (TPSA) and its higher number of atoms. This highly balanced flexibility is generally required for excellent and extreme positioning of the drug molecule to be more superimposable in the active binding pockets and cavities of any enzyme or protein (i.e., required for extreme lock-and-key positioning of the drug molecule after hitting and striking any protein molecule), leading to more adequate and potent inhibition (i.e., antagonism or blocking) of the actions (either catalytic or whatever) done or mediated by the protein. Consequently, the highly balanced flexibilities of CoViTris2020 and ChloViD2020 molecules greatly increase their abilities to be very potent anti-COVID-19 agents, respectively.
Collective screenshots of COVID-19 Docking Server outputs of the top predicted binding models resulted from the docking of CoViTris2020 molecule (colored pink) in different SARS-CoV-2 proteins (colored with other various colors; trace and cartoon styles), showing the extremely balanced high degrees of orientational and conformational flexibility of the molecule during the hitting attempts against all target coronaviral-2 proteins
Collective screenshots of COVID-19 Docking Server outputs of the top predicted binding models resulted from the docking of ChloViD2020 molecule (colored pink) in different SARS-CoV-2 proteins (colored with other various colors; trace and cartoon styles), showing the extremely balanced high degrees of orientational and conformational flexibility of the molecule during the hitting attempts against all target coronaviral-2 proteins
Remdesivir, computationally, comes first among the three reference drugs (and third among all tested compounds in general) in anti-COVID-19 activities with binding energies range of − 7.00 to − 9.10 kcal/mol. Remdesivir shows its best binding affinity with the target enzyme nsp14 (N7-MTase), as it binds to this important SARS-CoV-2 enzyme with a very good binding energy of − 9.10 kcal/mol. Ivermectin and favipiravir come last, with their best binding affinities observed with the two coronaviral-2 enzymes nsp14 (N7-MTase; − 7.20 kcal/mol) and RdRp (RTP site; − 6.90 kcal/mol), respectively.
The results of the inhibitory binding affinities of CoViTris2020 with the SARS-CoV-2 RdRp (RTP site) are relatively the best among all the COVID-19 targets, thus the next goal was to extensively investigate the specific interactions with the amino acids of the active site(s) of this crucial polymerase in deeper details (using the data files obtained from the same web server). These deep investigations revealed the high degree of similarity of the expected anti-RdRp mode of action of the two ligands CoViTris2020 and ChloViD2020 as compared to that of remdesivir and its active metabolite (some of the interacting residues of the active pockets are the same and some others are very close) as shown in Fig. 8a–c. It was also found that CoViTris2020 molecule has an apparent superiority over ChloViD2020 and remdesivir (in its active metabolic form) molecules (together with the other two reference molecules) in the strength of inhibitory binding forces with RdRp (e.g., CoViTris2020 has at least sixteen hydrogen bonds, more than four hydrophobic interactions, and at least one π-cation interaction; ChloViD2020 has at least nine hydrogen bonds, five hydrophobic interactions, and two halogen interactions; while on the other hand, remdesivir active metabolite (the most active drug among the three references) has only less than nine hydrogen bonds, no considerable hydrophobic interactions, and only one π-cation interaction), supporting its expected promising comprehensive anti-SAR-CoV-2 activities. Exceptionally, CoViTris2020 is the only inhibitor among all the investigated compounds that strongly interacts with the RdRp complex structure (especially chain A) with that large number of hydrogen bonds (of all types) which exceeds sixteen bonds. According to the previous literature, CoViTris2020 can be considered the only compound that strongly binds with the SARS-CoV-2 nsp12 in this extensively effective inhibitory mode, making it an optimal potent SARS-CoV-2 RdRp inhibitor candidate. Furthermore, this potential unique property gives CoViTris2020 an extra advantage of having potent blocking nature in its action on the active site of the polymerase. On the other hand, ChloViD2020 is the only inhibitor among all the investigated compounds that interacts with both chains A (nsp12) and C (nsp7) of the RdRp complex structure, since it, additionally, creates interaction forces of the hydrogen-bond/hydrophobic types with the polymerase cofactor chain C (mainly with the residues VAL12, SER15, GLN18, GLN19, and MET90). To the best of my knowledge, ChloViD2020 can be predictably considered one of the rarest ligands that interacts with and inhibits more than one protein chain of the SARS-CoV-2 RdRp complex (i.e., acts through a dual mode of action). ChloViD2020 is also characterized by its unique interaction forces created between its chlorine atom and the residues LYS417 and ASP418 of chain A. Table 2 summarizes all the main active amino acid residues involved in the inhibitory interactions of CoViTris2020, ChloViD2020, and remdesivir molecules, respectively.
In short, the previous computational results of the predicted binding modes of the two inhibitors CoViTris2020 and ChloViD2020 with the SARS-CoV-2/human proteins extremely comply with and support my suggested hypothetical multitarget mechanism of anti-COVID-19 action of each of the two ligands.
Anti-COVID-19 biological activities (in vitro assay) of CoViTris2020 and ChloViD2020
The results demonstrated in Table 3 obviously and directly revealed the extremely higher and amazing anti-COVID-19 effectiveness and potency of CoViTris2020 and ChloViD2020 (as two of the most potent anti-SARS-CoV-2 compounds ever) relative to those of each of the reference drugs. All the five tested compounds were found to inhibit SARS-CoV-2 replication in Vero E6 cells with EC50 under 100 μM. Surprisingly, CoViTris2020 (EC50 = 0.31 μM) was found to be about 65, 171, and 303.5 times more potent than remdesivir (EC50 = 20.17 μM), ivermectin (EC50 = 53.00 μM), and favipiravir (EC50 = 94.09 μM), respectively, in the anti-SARS-CoV-2 activity (in vitro). With the same interesting results, ChloViD2020 (EC50 = 1.01 μM) was found to be about 20, 52.5, and 93 times more potent than remdesivir, ivermectin, and favipiravir, respectively, in the same assay. According to this assay, CC50 of CoViTris2020 and ChloViD2020 is much larger than 100 μM, thus both compounds are expected to have very high safety margin and clinical selectivity index (SI; SI = CC50/EC50, SICoViTris2020 > 322.58 and SIChloViD2020 > 99.01), while on the other hand, the references remdesivir, ivermectin, and favipiravir are expected to have narrow safety margin and clinical therapeutic index (SIRemdesivir > 4.96, SIIvermectin > 1.89, and SIFavipiravir > 1.06). CoViTris2020 and ChloViD2020 are also having interestingly very small values of the concentration that causes 100% inhibition of the SARS-CoV-2 cytopathic effects in vitro (CoViTris2020 has the best CPEIC100 value, among all the tested compounds, of 0.99 μM, directly followed by ChloViD2020 which has CPEIC100 value of 1.97 μM) and of the concentration that is required for 50% reduction in the number of SARS-CoV-2 RNA copies in vitro (CoViTris2020 has the best EC50 value, among all the tested compounds, of 0.33 μM, directly followed by ChloViD2020 which has EC50 value of 1.23 μM).
We should put into consideration the possibility that CoViTris2020 and ChloViD2020 may undergo prior intracellular metabolic transformation into more active forms (e.g., their triphosphate forms) by human cellular enzymes (e.g., kinases and transferases), which may differ among various cell types, thus evaluation of the pharmacological actions of these two target compounds in primary human airway epithelial cells will clearly facilitate the interpretation and clarification of the previous results. The metabolic activation would almost add extra anti-COVID-19 activities to the two drugs through incorporation of human cell-biocompatible and human cell-bioavailable moieties into the chemical structures of both of them, and this would consequently increase the net clinical effectiveness and efficacy of these two potent compounds. Importantly, the three reference drugs (remdesivir, ivermectin, and favipiravir) are currently undergoing extensive and broad clinical trials as anti-SARS-CoV-2/anti-COVID-19 agents worldwide. The very high values of CC50 of CoViTris2020 and ChloViD2020 indicate that both compounds would be predictably well tolerated in the human body. The significantly desirable high values of SI (i.e., the extremely minute values of anti-SARS-CoV-2 EC50 along with the very high values of mammalian cells CC50) of CoViTris2020 and ChloViD2020 reflect that both compounds evidently favor resistant RNA virus over DNA virus and mammalian cells, and this, in turn, expectedly indicates and expresses the selective specificity of these two compounds as anti-COVID-19 drugs (specifically, CoViTris2020 can be considered as a unique superpowerful SARS-CoV-2 antidote/killer). CoViTris2020 is apparently more potent and more promising than ChloViD2020 as SARS-CoV-2 inhibitor. Using a combination formula of CoViTris2020 and remdesivir is a possible good experimental choice, as it may have exceptional and striking combinational synergistic anti-COVID-19 action (comprising the desirable advantages of both potent SARS-CoV-2 inhibitors) in further assays (in vivo) and preclinical/clinical trials. Almost all the factual experimental results obtained and concluded, here, in the anti-COVID-19 antiviral biological evaluation are complying with the previous speculative theoretical results suggested and extracted from the anti-SARS-CoV-2 computational molecular pharmacological predictions for the two newly repurposed biologically reevaluated compounds, CoViTris2020 and ChloViD2020, and the three reference drugs.
Conclusions
Thinking outside the box is very important to win our current challenge against the scary COVID-19 pandemic. Potent multiblockade of, mainly, the novel SARS-CoV-2 proteins including enzymes and receptors could be seen as one of the most effective fertile approaches for comprehensive COVID-19 therapy through designing, discovering, and searching for multitarget SARS-CoV-2 inhibitors, thus much efforts in drug discovery in 2020 were focused on trying to successfully repurpose known drugs and compounds in order to effectively inhibit this extremely resistant coronavirus. My specific efforts led to the discovery of two very promising potent multitarget SARS-CoV-2 inhibitors through the successful reevaluation and repurposing of the known antioxidant 1,3,4-oxadiazole compounds, previously synthesized by me, CoViTris2020 (1,2,3-tris[5-(3,4,5-trihydroxyphenyl)-1,3,4-oxadiazol-2-yl]propan-2-ol) and ChloViD2020 (5-[5-(7-chloro-4-hydroxyquinolin-3-yl)-1,3,4-oxadiazol-2-yl]benzene-1,2,3-triol), which effectively inhibited SARS-CoV-2 life cycle with IC50 values of 0.31 (according to the used anti-COVID-19 assay, CoViTris2020 is the most potent SARS-CoV-2 inhibitor discovered till now) and 1.01 μM, and amazingly presented about 65-/171-/303.5-fold and 20-/52.5-/93-fold anti-SARS-CoV-2 activities and potencies more than remdesivir/ivermectin/favipiravir, respectively. On the other hand, the discovery of the very potent anti-COVID-19 activities of CoViTris2020 and ChloViD2020 molecules through the successful biological reevaluation and repurposing opens the door for us to establish the first class of anti-COVID-19 agents of the type “polyphenolic 1,3,4-oxadiazoles” which will specifically comprise a series of 2,5-disubstituted-1,3,4-oxadiazole derivatives (beginning with the first two effective members, CoViTris2020 and ChloViD2020). Prior extensive computational molecular studies showed that CoViTris2020 and ChloViD2020 have the ideal and balanced values of the pharmacokinetic and druglikeness parameters required to be effectively potent anti-COVID-19 drugs inside the human body. Computational molecular modeling analysis of the best inhibitory docking binding modes of CoViTris2020 and ChloViD2020 molecules with the SARS-CoV-2/human proteins showed that the 3,4,5-trihydroxyphenyl moieties greatly increase the blocking affinities and potencies at the active and/or allosteric sites of the SARS-CoV-2/human enzymes or proteins (binding energies reach − 12.00 kcal/mol for CoViTris2020 and − 9.90 kcal/mol for ChloViD2020) when compared to those of the three references (which lack this effective antioxidant 3,4,5-trihydroxyphenyl moiety), remdesivir, ivermectin, and favipiravir (binding energies maximally reach − 9.10, − 7.20, and − 6.90 kcal/mol, respectively). Surprisingly, CoViTris2020 surpassed ChloViD2020 and the three moderately to highly potent reference drugs in the values of almost all compared computational and experimental anti-COVID-19 parameters, scores, and activities. Specifically, CoViTris2020 and ChloViD2020 give their best inhibitory binding affinities results with the catalytic active site of the SARS-CoV-2 RdRp. If the CoViTris2020 compound successfully passes the in vivo bioassays and then the preclinical/clinical trials with highly effective and satisfactorily significant results as anti-COVID-19 agent, a possible combination therapy (e.g., as an oral tablet/capsule, an intravenous/intramuscular parenteral vial/ampoule, or a nasal/oral/ocular prophylactic gel/drops) with a second highly potent old anti-RNA virus drug, such as remdesivir, may be a recommended available choice for ultimate COVID-19 treatment in the near future. In brief, in this new research paper, the antioxidant CoViTris2020 and ChloViD2020 molecules were successfully reevaluated, repurposed, and reported as very promising hit molecules (they could also be considered as the first extremely potent anticoronaviral-2 polyphenolic 1,3,4-oxadiazole compounds “Coronavirus-2 Killers”) with general multitarget and very potent successful inhibition against SARS-CoV-2 enzymes (mainly), and consequently, both compounds are two of the first known promising under-investigation candidate drugs for the effective and complete treatment of COVID-19.
Change history
03 September 2022
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s11030-022-10521-6
References
Hui DS, Azhar EI, Madani TA, Ntoumi F, Kock R, Dar O, Ippolito G, Mchugh TD, Memish ZA, Drosten C, Zumla A, Petersen E (2020) The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 91:264–266. https://doi.org/10.1016/j.ijid.2020.01.009
Li JY, You Z, Wang Q, Zhou ZJ, Qiu Y, Luo R, Ge XY (2020) The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future. Microbes Infect 22:80–85. https://doi.org/10.1016/j.micinf.2020.02.002
Jiang S, Du L, Shi Z (2020) An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg Microbes Infect 9:275–277. https://doi.org/10.1080/22221751.2020.1723441. Erratum: Jiang S (2020) Correction. Emerg Microbes Infect 9:539. https://doi.org/10.1080/22221751.2020.1737363
Dong L, Hu S, Gao J (2020) Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov Ther 14:58–60. https://doi.org/10.5582/ddt.2020.01012
Choy KT, Wong AY, Kaewpreedee P, Sia SF, Chen D, Hui KPY, Chu DKW, Chan MCW, Cheung PP, Huang X, Peiris M, Yen HL (2020) Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res 178:104786. https://doi.org/10.1016/j.antiviral.2020.104786
Elfiky AA (2020) Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci 248:117477. https://doi.org/10.1016/j.lfs.2020.117477
Elfiky AA (2020) Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci 253:117592. https://doi.org/10.1016/j.lfs.2020.117592. Erratum: Elfiky AA (2020) Corrigendum. Life Sci 258:118350. https://doi.org/10.1016/j.lfs.2020.118350
Shannon A, Le NT, Selisko B, Eydoux C, Alvarez K, Guillemot JC, Decroly E, Peersen O, Ferron F, Canard B (2020) Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 exonuclease active-sites. Antiviral Res 178:104793. https://doi.org/10.1016/j.antiviral.2020.104793
Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. https://doi.org/10.1038/s41422-020-0282-0
Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M, Chang S, Xie YC, Tian G, Jiang HW, Tao SC, Shen J, Jiang Y, Jiang H, Xu Y, Zhang S, Zhang Y, Xu HE (2020) Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368:1499–1504. https://doi.org/10.1126/science.abc1560
Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM (2020) The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res 178:104787. https://doi.org/10.1016/j.antiviral.2020.104787
Cai Q, Yang M, Liu D, Chen J, Shu D, Xia J, Liao X, Gu Y, Cai Q, Yang Y, Shen C, Li X, Peng L, Huang D, Zhang J, Zhang S, Wang F, Liu J, Chen L, Chen S, Wang Z, Zhang Z, Cao R, Zhong W, Liu Y, Liu L (2020) Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering 6:1192–1198. https://doi.org/10.1016/j.eng.2020.03.007
Du YX, Chen XP (2020) Favipiravir: pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin Pharmacol Ther 108:242–247. https://doi.org/10.1002/cpt.1844
Shiraki K, Daikoku T (2020) Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol Ther 209:107512. https://doi.org/10.1016/j.pharmthera.2020.107512
Derwand R, Scholz M (2020) Does zinc supplementation enhance the clinical efficacy of chloroquine/hydroxychloroquine to win today’s battle against COVID-19? Med Hypotheses 142:109815. https://doi.org/10.1016/j.mehy.2020.109815
Liu J, Cao R, Xu M, Wang X, Zhang H, Hu H, Li Y, Hu Z, Zhong W, Wang M (2020) Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov 6:16. https://doi.org/10.1038/s41421-020-0156-0
Wang X, Cao R, Zhang H, Liu J, Xu M, Hu H, Li Y, Zhao L, Li W, Sun X, Yang X, Shi Z, Deng F, Hu Z, Zhong W, Wang M (2020) The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov 6:28. https://doi.org/10.1038/s41421-020-0169-8
Abdul Amin S, Jha T (2020) Fight against novel coronavirus: a perspective of medicinal chemists. Eur J Med Chem 201:112559. https://doi.org/10.1016/j.ejmech.2020.112559
Goyal B, Goyal D (2020) Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb Sci 22:297–305. https://doi.org/10.1021/acscombsci.0c00058
Rabie AM, Tantawy AS, Badr SMI (2016) Design, synthesis, and biological evaluation of novel 5-substituted-2-(3,4,5-trihydroxyphenyl)-1,3,4-oxadiazoles as potent antioxidants. Am J Org Chem 6:54–80. https://doi.org/10.5923/j.ajoc.20160602.02
Ishida T (2019) Review on the role of Zn2+ ions in viral pathogenesis and the effect of Zn2+ ions for host cell-virus growth inhibition. Am J Biomed Sci Res 2:28–37. https://doi.org/10.34297/AJBSR.2019.02.000566
te Velthuis AJ, van den Worm SH, Sims AC, Baric RS, Snijder EJ, van Hemert MJ (2010) Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog 6:e1001176. https://doi.org/10.1371/journal.ppat.1001176
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25. https://doi.org/10.1016/S0169-409X(96)00423-1
Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H (2020) Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B 10:766–788. https://doi.org/10.1016/j.apsb.2020.02.008
COVID-19 Docking Server Web-based Software, COVID-19 Docking Server (Homepage on the Internet), Shan Change Lab., Institute of Bioinformatics and Medical Engineering, School of Electrical and Information Engineering, Jiangsu University of Technology, Changzhou 213001, China, 2020; Available from COVID-19 Docking Server on the Web (homepage: http://ncov.schanglab.org.cn); Results obtained through using interactive docking tool in COVID-19 Docking Server (Copyright© 2018–2023, Shan Chang; Version 2020) on this website (accessed and cited in 2020, 11–22 June), and references cited therein.
Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M (2015) PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res 43:W443–W447. https://doi.org/10.1093/nar/gkv315
Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM (2020) Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem 66:549–555. https://doi.org/10.1093/clinchem/hvaa029
Zhang L, Lin D, Kusov Y, Nian Y, Ma Q, Wang J, von Brunn A, Leyssen P, Lanko K, Neyts J, de Wilde A, Snijder EJ, Liu H, Hilgenfeld R (2020) α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J Med Chem 63:4562–4578. https://doi.org/10.1021/acs.jmedchem.9b01828
Acknowledgements
I gratefully thank and deeply acknowledge anyone who gave a hand to make this new discovery and work coming out to light.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
I hereby declare that I totally have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this new research paper.
Additional information
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s11030-022-10521-6
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rabie, A.M. RETRACTED ARTICLE: CoViTris2020 and ChloViD2020: a striking new hope in COVID-19 therapy. Mol Divers 25, 1839–1854 (2021). https://doi.org/10.1007/s11030-020-10169-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-020-10169-0