Extensive Wastewater-Based Epidemiology as a Resourceful Tool for SARS-CoV-2 Surveillance in a Low-to-Middle-Income Country through a Successful Collaborative Quest: WBE, Mobility, and Clinical Tests
Abstract
:1. Introduction
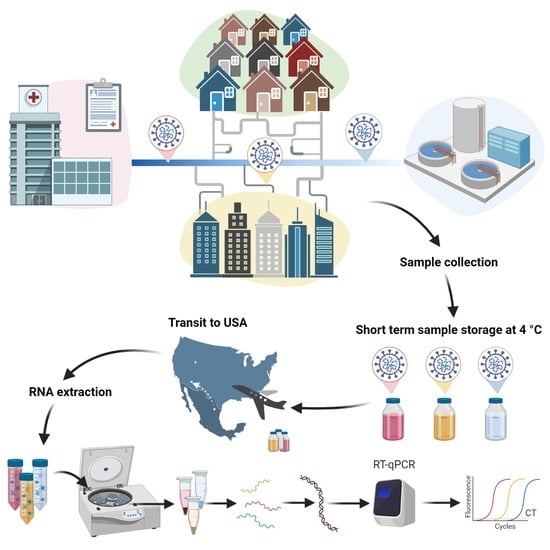
2. Materials and Methods
2.1. Study Area
2.2. Sample Collection, Storage, and Shipment
2.3. Sample Processing and Analysis
3. Results
3.1. SARS-CoV-2 in Wastewater in the Monterrey Metropolitan Area
3.2. COVID-19 Confirmed Clinical Cases vs. WW SARS-CoV-2 Viral Copies in the Monterrey Metropolitan Area
3.3. SARS-CoV-2 in WW and Mobility Info in the Monterrey Metropolitan Area
3.4. Limitations of the WBE Methodology in SARS-CoV-2 Detection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mexico: WHO Coronavirus Disease (COVID-19) Dashboard With Vaccination Data | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data n.d. Available online: https://covid19.who.int/region/amro/country/mx (accessed on 31 May 2022).
- Symptoms of COVID-19 | CDC n.d. Available online: https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html (accessed on 31 May 2022).
- Coronavirus Pandemic (COVID-19)—Our World in Data n.d. Available online: https://ourworldindata.org/coronavirus#citation (accessed on 31 May 2022).
- SCITEL n.d. Available online: https://www.inegi.org.mx/app/scitel/Default?ev=10 (accessed on 31 May 2022).
- Transporte de Pasajeros n.d. Available online: https://www.inegi.org.mx/temas/transporteurb/ (accessed on 31 May 2022).
- Resultados De La Encuesta Nacional De Ocupación Y Empleo. Nueva Edición (Enoen) Cifras Durante El Cuarto Trimestre De 2020 n.d. Available online: https://www.inegi.org.mx/contenidos/saladeprensa/boletines/2021/enoe_ie/enoe_ie2021_02.pdf (accessed on 31 May 2022).
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef] [PubMed]
- Panchal, D.; Prakash, O.; Bobde, P.; Pal, S. SARS-CoV-2: Sewage surveillance as an early warning system and challenges in developing countries. Environ. Sci. Pollut. Res. 2021, 28, 22221–22240. [Google Scholar] [CrossRef] [PubMed]
- Albert, S.; Ruíz, A.; Pemán, J.; Salavert, M.; Domingo-Calap, P. Lack of evidence for infectious SARS-CoV-2 in feces and sewage. Eur. J. Clin. Microbiol. 2021, 40, 2665–2667. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef] [PubMed]
- Rimoldi, S.G.; Stefani, F.; Gigantiello, A.; Polesello, S.; Comandatore, F.; Mileto, D.; Maresca, M.; Longobardi, C.; Mancon, A.; Romeri, F.; et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020, 744, 140911. [Google Scholar] [CrossRef]
- La Rosa, G.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Iaconelli, M.; Bonadonna, L.; Lucentini, L.; Suffredini, E. SARS-CoV-2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Sci. Total Environ. 2021, 750, 141711. [Google Scholar] [CrossRef]
- Westhaus, S.; Weber, F.-A.; Schiwy, S.; Linnemann, V.; Brinkmann, M.; Widera, M.; Greve, C.; Janke, A.; Hollert, H.; Wintgens, T.; et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany—Suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2020, 751, 141750. [Google Scholar] [CrossRef]
- Chavarria-miró, G.; Anfruns-Estrada, E.; Guix, S.; Paraira, M.; Galofré, B.; Sánchez, G.; Pintó, R.M.; Bosch, A. Sentinel Surveillance of SARS-CoV-2 in Wastewater Anticipates the Occurrence of SARS-CoV-2 Cases. medRxiv 2020. [Google Scholar] [CrossRef]
- Randazzo, W.; Cuevas-Ferrando, E.; Sanjuán, R.; Domingo-Calap, P.; Sánchez, G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health 2020, 230, 113621. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simón, P.; Allende, A.; Sánchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef]
- Kumar, M.; Patel, A.K.; Shah, A.V.; Raval, J.; Rajpara, N.; Joshi, M.; Joshi, C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020, 746, 141326. [Google Scholar] [CrossRef] [PubMed]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Green, H.; Wilder, M.; Collins, M.; Fenty, A.; Gentile, K.; Kmush, B.L.; Zeng, T.; Middleton, F.A.; Larsen, D.A. Quantification of SARS-CoV-2 and cross-assembly phage (crAssphage) from wastewater to monitor coronavirus transmission within communities. medRxiv 2020, 2, 1–18. [Google Scholar] [CrossRef]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Surya, K.; Buyukyoruk, M.; Cicha, C.; Vanderwood, K.K.; Wilkinson, R.; Wiedenheft, B. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. Cell Rep. Med. 2020, 1, 100098. [Google Scholar] [CrossRef]
- Peccia, J.; Zulli, A.; Brackney, D.E.; Grubaugh, N.D.; Kaplan, E.H.; Casanovas-Massana, A.; Ko, A.I.; Malik, A.A.; Wang, D.; Wang, M.; et al. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv 2020. [Google Scholar] [CrossRef]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci. Total. Environ. 2020, 743, 140621. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, J.; Xiao, A.; Gu, X.; Lee, W.L.; Armas, F.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N.; et al. SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases. mSystems 2020, 5, e00614-20. [Google Scholar] [CrossRef]
- Trottier, J.; Darques, R.; Mouheb, N.A.; Partiot, E.; Bakhache, W.; Deffieu, M.S.; Gaudin, R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health 2020, 10, 100157. [Google Scholar] [CrossRef]
- Wurtzer, S.; Marechal, V.; Mouchel, J.M.; Maday, Y.; Teyssou, R.; Richard, E.; Almayrac, J.L.; Moulin, L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv 2020. [Google Scholar] [CrossRef]
- Guerrero-Latorre, L.; Ballesteros, I.; Villacrés-Granda, I.; Granda, M.G.; Freire-Paspuel, B.; Ríos-Touma, B. SARS-CoV-2 in river water: Implications in low sanitation countries. Sci. Total Environ. 2020, 743, 140832. [Google Scholar] [CrossRef]
- Bar-Or, I.; Yaniv, K.; Shagan, M.; Ozer, E.; Weil, M.; Indenbaum, V.; Elul, M.; Erster, O.; Mendelson, E.; Mannasse, B.; et al. Regressing SARS-CoV-2 Sewage Measurements Onto COVID-19 Burden in the Population: A Proof-of-Concept for Quantitative Environmental Surveillance. Front. Public Health 2022, 9, 561710. [Google Scholar] [CrossRef] [PubMed]
- Kocamemi, B.A.; Kurt, H.; Hacıoglu, S.; Yaralı, C.; Saatci, A.M.; Pakdemirli, B. First data-set on SARS-CoV-2 detection for Istanbul wastewaters in Turkey. medRxiv 2020. [Google Scholar] [CrossRef]
- Haramoto, E.; Malla, B.; Thakali, O.; Kitajima, M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020, 737, 140405. [Google Scholar] [CrossRef]
- Fongaro, G.; Stoco, P.H.; Souza, D.S.M.; Grisard, E.C.; Magri, M.E.; Rogovski, P.; Schörner, M.A.; Barazzetti, F.H.; Christoff, A.P.; de Oliveira, L.F.V.; et al. The presence of SARS-CoV-2 RNA in human sewage in Santa Catarina, Brazil, November 2019. Sci. Total Environ. 2021, 778, 146198. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.T.; Rapp-Wright, H.; Egli, M.; Hartmann, A.; Steele, J.C.; Sosa-Hernández, J.E.; Melchor-Martínez, E.M.; Jacobs, M.; White, B.; Regan, F.; et al. High-throughput multi-residue quantification of contaminants of emerging concern in wastewaters enabled using direct injection liquid chromatography-tandem mass spectrometry. J. Hazard. Mater. 2020, 398, 122933. [Google Scholar] [CrossRef] [PubMed]
- Holland, L.A.; Kaelin, E.A.; Maqsood, R.; Estifanos, B.; Wu, L.I.; Varsani, A.; Halden, R.U.; Hogue, B.G.; Scotch, M.; Lim, E.S. An 81-Nucleotide Deletion in SARS-CoV-2 ORF7a Identified from Sentinel Surveillance in Arizona (January to March 2020). J. Virol. 2020, 94, e00711-20. [Google Scholar] [CrossRef] [PubMed]
- Bowes, D.A.; Driver, E.M.; Kraberger, S.; Fontenele, R.S.; Holland, L.A.; Wright, J.; Johnston, B.; Savic, S.; Newell, M.E.; Adhikari, S.; et al. Unrestricted Online Sharing of High-frequency, High-resolution Data on SARS-CoV-2 in Wastewater to Inform the COVID-19 Public Health Response in Greater Tempe, Arizona. medRxiv 2021. [Google Scholar] [CrossRef]
- COVID-19 Tablero México—CONACYT—CentroGeo—GeoInt—DataLab n.d. Available online: https://datos.covid-19.conacyt.mx/#DownZCSV (accessed on 31 May 2022).
- Rubio-Acero, R.; Beyerl, J.; Muenchhoff, M.; Roth, M.S.; Castelletti, N.; Paunovic, I.; Radon, K.; Springer, B.; Nagel, C.; Boehm, B.; et al. Spatially resolved qualified sewage spot sampling to track SARS-CoV-2 dynamics in Munich—One year of experience. Sci. Total Environ. 2021, 797, 149031. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Ronquillo, N.; Belda-Ferre, P.; Alvarado, D.; Javidi, T.; Longhurst, C.A.; Knight, R. High-Throughput Wastewater SARS-CoV-2 Detection Enables Forecasting of Community Infection Dynamics in San Diego County. mSystems 2021, 6, e00045-21. [Google Scholar] [CrossRef]
- Giraud-Billoud, M.; Cuervo, P.; Altamirano, J.C.; Pizarro, M.; Aranibar, J.N.; Catapano, A.; Cuello, H.; Masachessi, G.; Vega, I.A. Monitoring of SARS-CoV-2 RNA in wastewater as an epidemiological surveillance tool in Mendoza, Argentina. Sci. Total Environ. 2021, 796, 148887. [Google Scholar] [CrossRef]
- Carrillo-Reyes, J.; Barragán-Trinidad, M.; Buitrón, G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process Eng. 2020, 40, 101815. [Google Scholar] [CrossRef] [PubMed]
- Sala de Prensa IMSS. Gobierno de Mexico n.d. Available online: http://www.imss.gob.mx/prensa/archivo/202003 (accessed on 1 June 2022).
- Local mobility reports on COVID-19 n.d. Available online: https://www.google.com/covid19/mobility/ (accessed on 1 June 2022).
- Presentación de la “Actualización de medidas de mitigación contra COVID-19” | 02-07-2020 | Gobierno del Estado de Nuevo León n.d. Available online: https://www.nl.gob.mx/publicaciones/presentacion-de-la-actualizacion-de-medidas-de-mitigacion-contra-covid-19-02-07-2020 (accessed on 1 June 2022).
- Hasell, J.; Mathieu, E.; Beltekian, D.; Macdonald, B.; Giattino, C.; Ortiz-Ospina, E.; Roser, M.; Ritchie, H. A cross-country database of COVID-19 testing. Sci. Data 2020, 7, 345. [Google Scholar] [CrossRef] [PubMed]
- Balboa, S.; Mauricio-Iglesias, M.; Rodriguez, S.; Martínez-Lamas, L.; Vasallo, F.J.; Regueiro, B.; Lema, J.M. The fate of SARS-COV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021, 772, 145268. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sosa-Hernández, J.E.; Oyervides-Muñoz, M.A.; Melchor-Martínez, E.M.; Driver, E.M.; Bowes, D.A.; Kraberger, S.; Lucero-Saucedo, S.L.; Fontenele, R.S.; Parra-Arroyo, L.; Holland, L.A.; et al. Extensive Wastewater-Based Epidemiology as a Resourceful Tool for SARS-CoV-2 Surveillance in a Low-to-Middle-Income Country through a Successful Collaborative Quest: WBE, Mobility, and Clinical Tests. Water 2022, 14, 1842. https://doi.org/10.3390/w14121842
Sosa-Hernández JE, Oyervides-Muñoz MA, Melchor-Martínez EM, Driver EM, Bowes DA, Kraberger S, Lucero-Saucedo SL, Fontenele RS, Parra-Arroyo L, Holland LA, et al. Extensive Wastewater-Based Epidemiology as a Resourceful Tool for SARS-CoV-2 Surveillance in a Low-to-Middle-Income Country through a Successful Collaborative Quest: WBE, Mobility, and Clinical Tests. Water. 2022; 14(12):1842. https://doi.org/10.3390/w14121842
Chicago/Turabian StyleSosa-Hernández, Juan Eduardo, Mariel Araceli Oyervides-Muñoz, Elda M. Melchor-Martínez, Erin M. Driver, Devin A. Bowes, Simona Kraberger, Sofia Liliana Lucero-Saucedo, Rafaela S. Fontenele, Lizeth Parra-Arroyo, LaRinda A. Holland, and et al. 2022. "Extensive Wastewater-Based Epidemiology as a Resourceful Tool for SARS-CoV-2 Surveillance in a Low-to-Middle-Income Country through a Successful Collaborative Quest: WBE, Mobility, and Clinical Tests" Water 14, no. 12: 1842. https://doi.org/10.3390/w14121842