Interplay between swine enteric coronaviruses and host innate immune
- State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, China
Swine enteric coronavirus (SeCoV) causes acute diarrhea, vomiting, dehydration, and high mortality in neonatal piglets, causing severe losses worldwide. SeCoV includes the following four members: transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), porcine delta coronavirus (PDCoV), and swine acute diarrhea syndrome coronavirus (SADS-CoV). Clinically, mixed infections with several SeCoVs, which are more common in global farms, cause widespread infections. It is worth noting that PDCoV has a broader host range, suggesting the risk of PDCoV transmission across species, posing a serious threat to public health and global security. Studies have begun to focus on investigating the interaction between SeCoV and its host. Here, we summarize the effects of viral proteins on apoptosis, autophagy, and innate immunity induced by SeCoV, providing a theoretical basis for an in-depth understanding of the pathogenic mechanism of coronavirus.
1. Introduction
1.1. Swine enteric coronavirus
Coronaviruses are one of the most devastating pathogens. The sudden outbreak of COVID-19 in 2019 has had a major impact on global public health and economic development, while the devastating effects of CoVs are not only limited to humans but also occur in livestock populations. Swine enteric coronaviruses (SeCoV) pose a huge threat to the global farm industry. Four SeCoVs were identified: porcine epidemic diarrhea virus (PEDV), porcine transmissible gastroenteritis virus (TGEV), porcine delta coronavirus (PDCoV), and swine acute diarrhea syndrome coronavirus (SADS-CoV). In the last century, PEDV and TGEV were first reported (1–3), and then widely spread to many swine-producing countries in Europe and Asia (4, 5). Recently, PDCoV and SADS-CoV have emerged as SeCoVs (6, 7), Compared with PEDV and TGEV, the clinical signs caused by PDCoV and SADS-CoV infection are less severe, and the mortality rate of newborn piglets is 30–40%. It is worth mentioning that in 2021, US scientists discovered that the plasma samples of three Haitian children with unexplained fever tested positive for PDCoV, in which suggesting the risk of PDCoV across species transmission (8).
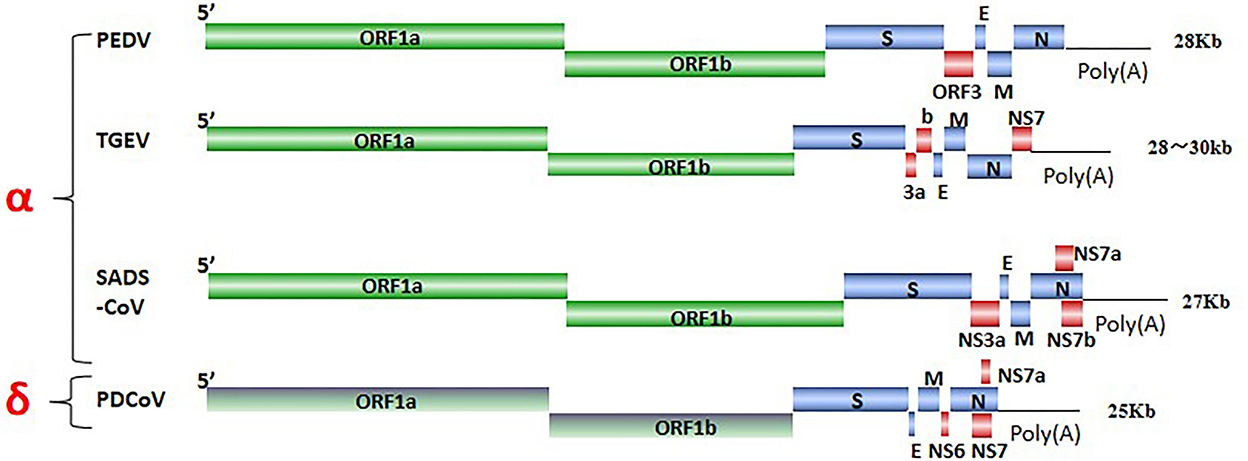
SeCoV is a single-stranded, positive-sense RNA virus, and its viral genome consists of structural proteins, non-structural proteins, and accessory proteins. Four structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins were identified. The S protein mediates attachment to the host receptor and is a trimer with an S1 subunit that contains the large receptor-binding domain (RBD) and an S2 subunit that contains peptides mediating cell fusion (9). E and M proteins are responsible for maintaining the structure and size of the viral envelope (10). The N protein constitutes the only protein present in the nucleocapsid and wraps the virus genome to form a nucleoprotein complex (11). ORF1ab encodes non-structural proteins via nsp3 and nsp5 cleavage, there are 15–16 functional non-structural proteins (nsps), These nsps are involved in the replication and transcription of viral RNA and some nsps also inhibit the host immune response (10, 12), ORF3, NS6, and NS7 encode accessory proteins to modulate viral pathogenicity (13) (Figure 1). SeCoV has become a major cause of lethal watery diarrhea in newborn piglets, imposing enormous economic losses and a public health burden on the swine industry worldwide.
1.2. Autophagy induce by SeCoV infection
Autophagy is a process in which cells use lysosomes to degrade damaged organelles and macromolecular substances under the regulation of autophagy-related genes (Atg). Previous studies have reported that autophagy is an intrinsic host defense mechanism that mediates the autophagic elimination of viral constituents or virions by targeting virus particles or virus component degradation to facilitate host innate and adaptive immunity. Increasing evidence indicates that viruses have evolved various complex strategies to escape or subvert the antiviral effects of autophagy (Figure 2). For example, SARS-CoV-2 ORF3a promotes the induction of autophagy via the classic ATF6 and IRE1-XBP1 UPR pathways to protect the virus from hydrolysis (14). Furthermore, ORF10 and M of SARS-CoV-2 promote the accumulation of LC3 in mitochondria and induce mitophagy, which inhibits RIG-MAVS-triggered IFN signaling (15, 16). PEDV-triggered autophagy in Vero cells via both the PERK and IER1 pathways promotes viral replication (17–19). Additionally, nsp6 and ORF3 of PEDV were able to induce significant autophagy in IPEC-J2 cells, and nsp6 of PEDV induced autophagy by inhibiting the PI3K/Akt/mTOR signaling pathway, which promotes cell damage and enhances the virulence of PEDV (20). Moreover, PEDV ORF3 protein triggers the endoplasmic reticulum (ER) stress response by upregulating the expression of GRP78 protein and activating the PERK-eIF2α signaling pathway to induce autophagy (21). In addition, SADS-CoV and PDCoV induce autophagy to facilitate viral replication via the PI3K/Akt/mTOR signaling pathway in vitro (22–24).
TGEV infection induces mitophagy to suppress oxidative stress and apoptosis in porcine epithelial cells (IPEC-J2 cells) to promote cell survival and, possibly, viral infection. Furthermore, N of TGEV may be involved in mitochondrial damage and mitophagy induction during TGEV infection (25). Interestingly, TGEV infection activates autophagy, whereas autophagy inhibits TGEV replication (26). Upon PDCoV infection, the upregulation of the LC3-II/LC3-I ratio and the downregulation of p62 protein levels indicate that PDCoV infection may induce autophagy, similar to other CoVs (22, 27). Additionally, PDCoV-induced autophagy enhances viral replication through the p38 signaling pathway (28).
1.3. Apoptosis induce by SeCoV infection
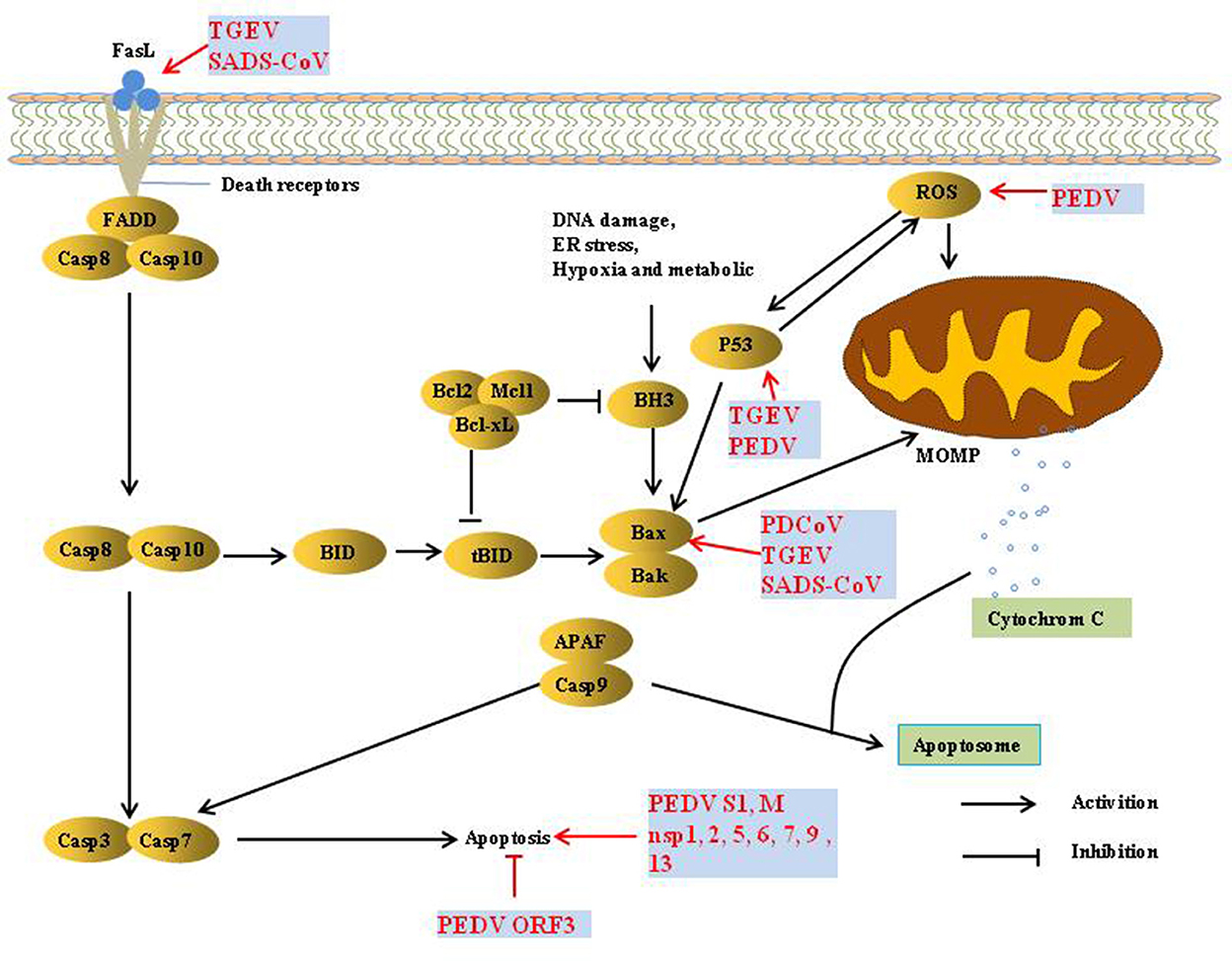
Apoptosis refers to the autonomous and orderly death of cells that is controlled by genes to maintain the stability of the internal environment. It is involved in the activation, expression, and regulation of a series of genes. There are two pathways of apoptosis, extrinsic and intrinsic. The extrinsic apoptotic pathway is mediated by death receptors (DRs) on the cell membrane, thereby activating the cascade of apoptosis signaling pathways. The intrinsic apoptosis pathway is mainly activated by apoptosis inducers in the cytoplasm to activate mitochondrial pro-apoptotic factors and destroy the integrity of the mitochondrial outer membrane. Subsequently, the increase in mitochondrial outer membrane permeability (MOMP) promotes the release of cytochrome c (Cyt c) and apoptosis-inducing factor (AIF), thereby inducing apoptosis (29).
Apoptosis is considered a host innate defense mechanism that disrupts viral replication by eliminating virus-infected cells, but some viruses utilize apoptosis as a mechanism for cell killing and viral spread (30) (Figure 3). PEDV infection induces apoptosis via a caspase-independent mitochondrial AIF-mediated pathway to facilitate viral replication (31–33). It has been demonstrated that the activation of p38 MAPK and JNK cascades also contributes to PEDV replication, but they are not linked to PEDV-mediated apoptosis (34, 35). P53 plays an essential role in viral infection-induced apoptosis. PEDV and TGEV induce apoptosis via a P53-dependent pathway (35, 36). PEDV infection activated P53-puma and reactive oxygen species (ROS)/p53 signaling pathways to induce apoptosis in Vero cells and cause cell cycle arrest at the G0/G1 phase (35, 37). The S1 protein of many coronaviruses can induce cell apoptosis; the PEDV S1 protein is the main inducer of cell apoptosis during PEDV infection, and PEDV M and nsp1, 2, 5, 6, 7, 9, and 13 also induce cell apoptosis, but to a lesser extent. Similarly, the S protein of TGEV can strongly induce apoptosis in Vero-E6 cells, suggesting that S1 is a promising strategy to inhibit coronavirus infection (38, 39). In contrast, reverse genetics technology has been used to prove that the PEDV ORF3 protein promotes virus proliferation by inhibiting cell apoptosis (40).
TGEV induced apoptosis in PK-15 and ST cells but not in intestinal epithelial cells (41, 42), p53- and ROS-mediated AIF pathways, and caspase-dependent pathways both played a dominant role in triggering apoptosis. However, p38 MAPK signaling was only partially responsible for the activation of p53 and contributed less to TGEV-induced apoptosis (36, 42–45). Interestingly, the TGEV N protein is cleaved by caspase-6 and−7 during TGEV-induced apoptosis (46). However, TGEV N upregulated p53 and p21 and arrested the cell cycle at the S and G2/M phases, finally resulting in apoptosis of PK-15 cells (47).
PDCoV induces apoptosis to promote viral replication in both LLC-PK and ST cells but not in infected intestinal enterocytes in vivo (48). In addition, PDCoV and SADS-CoV infection induces apoptosis by recruiting Bax or opening the mitochondrial permeability transition pore (MPTP) and then releasing Cyt c, sequentially activating initiator caspase-9 and downstream effector caspase-3, thereby orchestrating the final apoptotic response to facilitate viral replication in vitro, Intrinsic caspase-9 dependent apoptosis pathway plays an important role in the successful replication of PDCoV and SADS-CoV (49, 50). Further studies showed that caspase-dependent FASL-mediated apoptotic pathways are also involved in SADS-CoV infection (50).
1.4. Innate immunity recognition of SeCoV
1.4.1. Pattern recognition receptors
The innate immune response is the host's first line of defense against pathogens. Innate immune cells recognize pathogen-associated molecular patterns (PAMP) by expressing pattern-recognition receptors (PRRs), including Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), NOD-like receptors (NLRs), AIM2-like receptors (ALRs), C-type lectin receptors (CLRs), and intracellular DNA sensors, such as cyclic GMP-AMP synthase (cGAS), which are key innate immune components that recognize viral components such as viral nucleic acids and proteins (51). Among these receptors, TLRs and RLRs are the two major receptors responsible for sensing RNA virus infections and triggering antiviral IFN programs. The TLR family comprises 10 members (TLR1–TLR10) in humans and 12 (TLR1–TLR9 and TLR11–TLR13) in mice (52). TLR1, TLR2, TLR4, TLR5, and TLR6 play pivotal roles in viral protein recognition (53). The membrane proteins TLR3, TLR7/8, and TLR9 are used, respectively (54–56). The RLR family includes three members: retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2). RIG-I and MDA5 are activated by immunostimulatory RNA, which leads to the activation of cytoplasmic kinases that promote the activation of interferon regulatory factor 3 (IRF3), IRF7, and nuclear factor-kappa B (NF-κB). Activated IRF3/IRF7 binds to PRD I/III sequences and induces the expression of type I IFN genes (57). The activated form of NF-κB translocates to the nucleus and triggers IFN-β expression by binding to PRD II elements (58). IFN is then secreted, which binds to receptors on virus-infected cells, as well as uninfected neighboring cells, and activates the JAK/STAT pathway to generate hundreds of ISGs to establish an antiviral state (59).
1.4.2. Immune evasion mechanisms of SeCoV
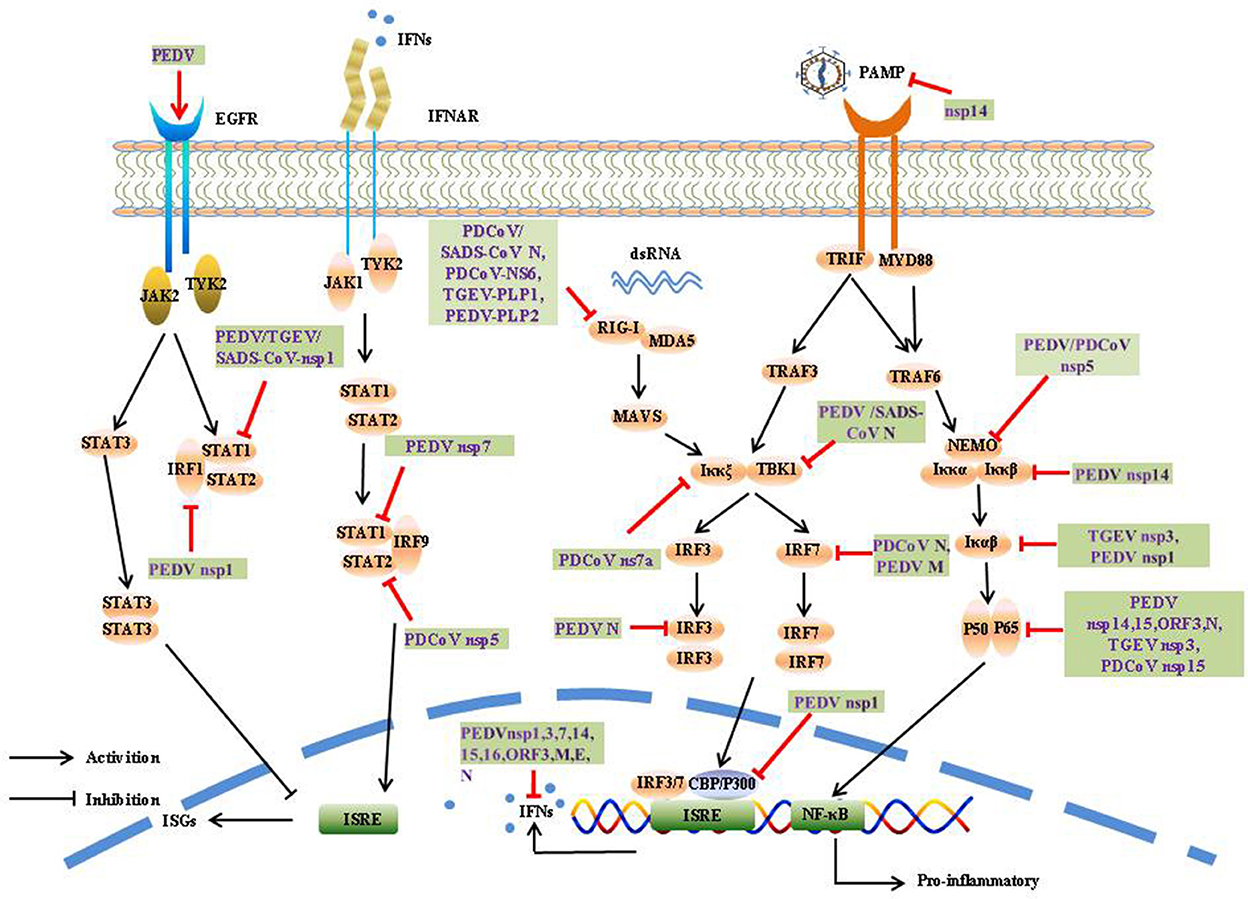
The induction of IFN-α/β is the most rapid and effective mechanism by which the host initiates innate immune responses. To counter innate immune signaling, many coronaviruses have evolved different strategies to develop multiple strategies to evade the innate immune response and efficiently promote their replication and infective capacity, particularly by minimizing IFN production and inhibiting IFN signaling (Figure 4).
1.4.2.1. Ubiquitination and deubiquitination induced by SeCoV
Ubiquitination is a critical biological process in the post-translational modification of proteins and involves multiple signaling pathways such as protein metabolism, apoptosis, DNA damage, cell-cycle progression, and cancer development. Ubiquitin is mainly connected in eight ways (M1, K6, K11, K27, K29, K33, K48, and K63), and can regulate different functions of substrate proteins. For example, K48 polyubiquitination mainly plays the role of the ubiquitin-proteasome system to degrade substrates and proteins, whereas K63 polyubiquitination mainly regulates endocytosis, protein interaction, and signal transduction (60).
The CoV N protein, the most abundant viral protein, plays a key role in IFN interruption. The SADS-CoV N protein mediates K27-, K48-, and K63-linked ubiquitination of RIG-I and its subsequent proteasome-dependent degradation to inhibit the host IFN response (61). The PDCoV N protein could directly target porcine RIG-I to interfere with its binding to dsRNA and block its early activation by blocking porcine Riplet (pRiplet)-mediated K63-linked polyubiquitination, thus suppressing IFN-β Production (62). In addition, PDCoV N protein promotes poIRF7 degradation through the K6, K11, and K29 polyubiquitination-proteasome pathways to reduce type-I IFN production (63).
The deubiquitinase (DUB) family is responsible for the specific hydrolysis of ubiquitin molecules from ubiquitin-linked proteins or precursor proteolysis, which affects the localization, stability, and function of target proteins in cells, DUB are widely present in various viruses, and significantly influences viral activity. Interestingly, all CoV DUB activities are mediated by PLPs, and the PLPs of human coronavirus NL63 (HCoV-NL63), SARS-CoV, MHV, and MERS-CoV significantly reduced the levels of ubiquitinated STING, RIG-I, TBK1, and IRF-3, thereby negatively affecting the regulation of host antiviral innate immunity (64). Likewise, PEDV PLP2 and TGEV PL1 strongly inhibit RIG-I- and STING-activated IFN expression via deubiquitination (65, 66). A recent study showed that SADS-CoV PLP2 could also function as a DUB, such as PEDV PLP2, SARS-CoV PLpro, and TGEV PLP1 (67).
1.4.2.2. Protein cleavage
SeCoV encodes non-structural protein 5, also called the 3C-like protease, and is responsible for coronavirus polyprotein processing. It cleaves the polyprotein at more than 11 sites to yield the essential proteins required for virus replication and pathogenesis. At the same time, the protease can also use its cleavage activity to cleave host proteins, especially the key molecules of IFN production and signal transduction, and play an immunomodulatory role. 3CLpro is an attractive drug target because it is highly conserved among known coronavirus species. Many viruses antagonize innate immune signaling by cleaving 3C-like proteases; for example, porcine sapelovirus (PSV) 3Cpro inhibits the production of IFN-β by cleaving MAVS and degrading MDA5 and TBK1 (68). Enterovirus 71 (EV71) 3C interacts with and cleaves TAB2 and TAK1 to interfere with the inflammatory responses (69). Norovirus (NoV) encoding a 3C-like protease was found to effectively suppress Sendai virus (SEV)-mediated IFN-β production by cleaving the NF-κB essential modulator (NEMO) (70). Similar to NoV and EV71 3Cpro, PEDV and PDCoV encode a 3C-like protease, nsp5, which is an IFN antagonist that cleaves NEMO at Q231, suggesting that NEMO may be a common target for coronaviruses (71, 72). In addition, PDCoV nsp5 also suppressed IFN signaling by cleaving STAT2, a key molecule in the JAK-STAT pathway, nsp5 cleaved STAT2 at both Q685 and Q758 (73).
1.4.2.3. Competitive binding
It has been demonstrated PEDV N protein directly interacts with TBK1, thereby sequestering the association between TBK1 and IRF3, which in turn inhibits both IRF3 activation and type I IFN production (74). Moreover, the PDCoV N protein antagonizes IFN-β production by interfering with the binding of dsRNA and protein activator of protein kinase R (PACT) to RIG-I (75). The SADS-CoV N protein suppresses the RLRs Signaling pathway. Moreover, the SADS-CoV N Protein not only blocked the IPS-1–TBK1 interaction but also disrupted the formation of the TNF receptor-associated factor 3(TRAF3)–TBK1 complex, which led to reduced TBK1 activation and IFN-β production (76). It has been demonstrated PEDV nsp7 antagonizes type I IFN production, PEDV nsp7 also antagonized IFN-α-induced JAK-STAT signaling by sequestering the interaction between karyopherin α1 (KPNA1) and STAT1 (77). Cytoplasmic stress granules (SGs) can effectively exert antiviral functions; however, nsp15s of PEDV, TGEV, SARS-CoV, and SARS-CoV-2 have conserved functions that interfere with chemically induced SGs formation (78). Coronavirus accessory proteins are species specific and have low homology with other known proteins. Although current research has shown that coronavirus accessory proteins are not necessary for virus replication (79), extensive reports have indicated that many accessory proteins are involved in immune regulation and virus virulence. For example, PDCoV NS6 interacts with the CTD of RIG-I and the Hel and CTD of MDA5, and this interaction attenuates the binding of RIG-I/MDA5-dsRNA, resulting in a reduction in IFN-β production (80). PDCoV NS7a interacts with IKKε, which significantly disrupts the interaction between IKKε and TRAF3 or IRF3, thereby inhibiting IFN-β production (81).
1.4.2.4. Impair phosphorylation or suppressed the nuclear translocation
Post-translational modification of proteins is a critical way to regulate protein function. Phosphorylation is one of the most extensively investigated post-translational modifications involved in the regulation of signal transduction, but viral encoded proteins can regulate phosphorylation and dephosphorylation to promote proliferation. The PEDV E protein has been found to block the production of IFN, but little is known about the process by which the E protein subverts host innate immunity. A previous study showed that PEDV E protein is responsible for inducing ER stress through activation of the PERK/eIF2α branch and activation of NF-κB (82). Further studies showed that PEDV E protein remarkably suppressed IFN-β production by interfering with the translocation of IRF3 from the cytoplasm to the nucleus through direct interaction with IRF3 (83, 84). PEDV N protein blocks NF-κB nuclear translocation to antagonize IFN-λ production (85). The PEDV M protein plays an important role in viral assembly, viral budding, and host immune mediation. PEDV M protein interacts with IRF7 and significantly suppresses its phosphorylation and dimerization of IRF7, leading to decreased expression of type I IFN (86). Existing study has been identified the sole accessory protein ORF3 of PEDV as NF-κB antagonist, it inhibits the phosphorylation of IκBα, in addition, PEDV ORF3 inhibits NF-κB activation by interfering the phosphorylation and expression of p65, as well as interfering nuclear translocation of p65, which ultimately leaded to the inhibition of IL-6 and IL-8 production (87). As a key virulence factor for coronaviruses, nsp1 impedes host protein expression via multiple mechanisms. Of the 21 PEDV proteins, nsp1, nsp3, nsp5, nsp7, nsp14, nsp15, nsp16, ORF3, and E inhibited NF-κB activity, and nsp1 appeared to be the most potent inhibitor. Nsp1 interfered with the phosphorylation and degradation of IκBα, and thus blocked p65 nuclear transport; however, PEDV nsp1 did not interfere with IRF3 phosphorylation and nuclear translocation, which interrupted the enhanceosome assembly of IRF3 and CREB-binding protein (CBP) by degrading CBP, resulting in the inhibition of ISGs expression (88, 89). Furthermore, nsp1 was found to suppress type III IFN activity by blocking the nuclear translocation of interferon regulatory factor 1 (IRF1) and reducing the number of peroxisomes (90). PEDV, TGEV, and SARS-CoV nsp1 significantly inhibited the phosphorylation of STAT1 at S727, interfering with the effect of IFN-I, and nsp1 also arrested host cells to stay in the G0/G1 phase (91, 92). In contrast, PEDV nsp1 inhibited CCAAT/enhancer-binding protein β (C/EBP-β) phosphorylation to reduce complement component 3 (C3) expression, which is considered to play a crucial role in preventing viral infection (93). Nsp14 of CoV has ExoN and guanine-N7-methyltransferase (N7-MTase) activities (94, 95), playing a key role in viral mRNA cap synthesis, CoV replication and transcription, Howerver, the function and mechanism by which nsp14 modulates and manipulates host immune responses remain largely unknown, Recently study showed PEDV nsp14 remarkably decreased NF-κB activation and proinflammatory cytokines expression, it interacted with Iκκs and p65 to inhibite the phosphorylation of Iκκs. Furthermore, nsp14 suppresses TNF-α-induced phosphorylation and nuclear import of p65 (96). TGEV nsp3 has been shown to strongly inhibits NF-κB signaling by suppressing Iκβα degradation and inhibiting p65 phosphorylation and nuclear translocation (97). Nsp15 encodes an endoribonuclease that conserves all coronaviruses. The nuclease activity of nsp15 plays a critical role in viral evasion by triggering an innate immune response. PDCoV nsp15 significantly inhibits IFN-β production by disrupting the phosphorylation and nuclear translocation of p65, independent of its endoribonuclease (98). TGEV ORF7 binds to the catalytic subunit of protein phosphatase 1 (PP1c) and regulated the dephosphorylation of eIF2α to counteract host cellular defenses (99). In addition, deletion of ORF7 increased innate immune responses and acute tissue damage, demonstrating antagonism from the opposite perspective (100).
1.4.2.5. Degradation and inactivation induced by SeCoV
In addition, SeCoV can antagonize the host innate immune response through degradation and inactivation. PEDV suppresses type I interferon response by stimulating epidermal growth factor receptor (EGFR) activation, which is responsible for STAT3 expression (101). PEDV nsp15 directly degrades the mRNA of TBK1 and IRF3 depending on its EndoU activity to inhibit the production of IFN and ISG and antagonize the host innate response to promote replication (102). CoV nsp14 can degrade dsRNA PAMPs to prevent IFN induction during CoV infection (103). Of the 21 PEDV proteins, nsp1, nsp3, nsp7, nsp14, nsp15, and nsp16 were found to inhibit IFN-β and IRF3 promoter activity (89). Further studies showed that nsp1, nsp3, nsp5, nsp8, nsp14, nsp15, nsp16, ORF3, E, M, and N suppressed type III IFN activity (90).
2. Discussion
SeCoV is a pathogenic microorganism that seriously threatens the pig industry and causes massive economic loss. The above evidence reveals the viral immune evasion mechanisms of SeCoV, where the origin of SeCoV and the interaction between the virus and host need to be further elucidated. Furthermore, the rapid global spread of highly pathogenic of SARS-CoV, MERS-CoV, and SADS-CoV-2 pose a concern about cross-species transmission, such as the discovery of PDCoV in Haitian children. It is evident that proper surveillance of viral biodiversity can be used to prevent animals becoming mixers and intermediate hosts of various coronaviruses in the future. Morever, an important feature of the epidemiology of SECoV is the emergence of several different variants, which vary in their transmissibility, virulence, clinical disease presentation, and vaccines response, resulting in unforeseeable epidemic scope and pathogenicity. Up to now, porcine aminopeptidase N (pAPN) has been identified as a receptor for TGEV, but the receptors of PEDV, PDCoV, and SASD-CoV remain unknown, hindering the development of vaccines and drugs.
Exploration of these programs will help us further understand how SeCoV exists to ensure their survival, and also provide us with new ideas for developing drug targets for the prevention and treatment of SeCoV.
Author contributions
ML wrote the first draft of the manuscript. LG and LF contributed to conception and design of the review. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
The manuscript was supported by the National Key R&D Program of China (2021YFD1801105), Natural Science Foundation of Heilongjiang Province of China (YQ2020C023), and National Natural Science Foundation of China (31872474).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wood EN. An apparently new syndrome of porcine epidemic diarrhoea. Vet Rec. (1977) 100:243–4. doi: 10.1136/vr.100.12.243
2. Doyle LP, Hutchings LM. A Transmissible gastroenteritis in pigs. J Am Vet Med Assoc. (1946) 108:257–9.
3. Pensaert MB, Bouck PD. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. (1978) 58:243–7. doi: 10.1007/BF01317606
4. Hou Y, Yue X, Cai X, Wang S, Liu Y, Yuan C, et al. Complete genome of transmissible gastroenteritis virus ayu strain isolated in Shanghai, China. J Virol. (2012) 86:11935. doi: 10.1128/JVI.01839-12
5. Weiwei H, Qinghua Y, Liqi Z, Haofei L, Shanshan Z, Qi G, et al. Complete genomic sequence of the coronavirus transmissible gastroenteritis virus shxb isolated in China. Arch Virol. (2014) 159:2295–302. doi: 10.1007/s00705-014-2080-9
6. Woo PCY, Lau SKP, Lam CSF, Lau CCY, Tsang AKL, Lau JHN, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. (2012) 86:3995–4008. doi: 10.1128/JVI.06540-11
7. Pan Y, Tian X, Qin P, Wang B, Zhao P, Yang YL, et al. Discovery of a novel swine enteric alphacoronavirus (Seacov) in Southern China. Vet Microbiol. (2017) 211:15–21. doi: 10.1016/j.vetmic.2017.09.020
8. Lednicky JA, Tagliamonte MS, White SK, Elbadry MA, Alam MM, Stephenson CJ, et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature. (2021) 600:133–7. doi: 10.1038/s41586-021-04111-z
9. de Groot RJ, Luytjes W, Horzinek MC, van der Zeijst BA, Spaan WJ, Lenstra JA. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J Mol Biol. (1987) 196:963–6. doi: 10.1016/0022-2836(87)90422-0
10. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. (2015) 1282:1–23. doi: 10.1007/978-1-4939-2438-7_1
11. Masters PS. The molecular biology of coronaviruses. Adv Virus Res. (2006) 66:193–292. doi: 10.1016/S0065-3527(06)66005-3
12. Zhang J. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. (2016) 226:71–84. doi: 10.1016/j.virusres.2016.05.028
13. Koonpaew S, Teeravechyan S, Frantz PN, Chailangkarn T, Jongkaewwattana A. Pedv and Pdcov pathogenesis: the interplay between host innate immune responses and porcine enteric coronaviruses. Front Vet Sci. (2019) 6:34. doi: 10.3389/fvets.2019.00034
14. Su WQ, Yu XJ, Zhou CM. SARS-CoV-2 ORF3a induces incomplete autophagy via the unfolded protein response. Viruses. (2021) 13:2467. doi: 10.3390/v13122467
15. Hui X, Zhang L, Cao L, Huang K, Zhao Y, Zhang Y, et al. SARS-CoV-2 promote autophagy to suppress type i interferon response. Signal Transd Targeted Ther. (2021) 6:180. doi: 10.1038/s41392-021-00574-8
16. Li X, Hou P, Ma W, Wang X, Wang H, Yu Z, et al. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading mavs through mitophagy. Cell Mol Immunol. (2022) 19:67–78. doi: 10.1038/s41423-021-00807-4
17. Guo X, Zhang M, Zhang X, Tan X, Guo H, Zeng W, et al. Porcine epidemic diarrhea virus induces autophagy to benefit its replication. Viruses. (2017) 9:53. doi: 10.3390/v9030053
18. Sun P, Jin J, Wang L, Wang J, Zhou H, Zhang Q, et al. Porcine epidemic diarrhea virus infections induce autophagy in vero cells via ros-dependent endoplasmic reticulum stress through Perk and Ire1 Pathways. Vet Microbiol. (2021) 253:108959. doi: 10.1016/j.vetmic.2020.108959
19. Park JY, Ryu J, Hong EJ, Shin HJ. Porcine epidemic diarrhea virus infection induces autophagosome formation but inhibits autolysosome formation during replication. Viruses. (2022) 14:1050. doi: 10.3390/v14051050
20. Lin H, Li B, Liu M, Zhou H, He K, Fan H. Nonstructural protein 6 of porcine epidemic diarrhea virus induces autophagy to promote viral replication via the Pi3k/Akt/Mtor axis. Vet Microbiol. (2020) 244:108684. doi: 10.1016/j.vetmic.2020.108684
21. Zou D, Xu J, Duan X, Xu X, Li P, Cheng L, et al. Porcine epidemic diarrhea virus Orf3 protein causes endoplasmic reticulum stress to facilitate autophagy. Vet Microbiol. (2019) 235:209–19. doi: 10.1016/j.vetmic.2019.07.005
22. Qin P, Du EZ, Luo WT, Yang YL, Zhang YQ, Wang B, et al. Characteristics of the life cycle of porcine deltacoronavirus (Pdcov) in vitro: replication kinetics, cellular ultrastructure and virion morphology, and evidence of inducing autophagy. Viruses. (2019) 11:455. doi: 10.3390/v11050455
23. Zeng S, Peng O, Sun R, Xu Q, Hu F, Zhao Y, et al. Transcriptional landscape of Vero E6 cells during early swine acute diarrhea syndrome coronavirus infection. Viruses. (2021) 13:674. doi: 10.3390/v13040674
24. Zhou X, Zhou L, Ge X, Guo X, Han J, Zhang Y, et al. Quantitative proteomic analysis of porcine intestinal epithelial cells infected with porcine deltacoronavirus using itraq-coupled Lc-Ms/Ms. J Proteome Res. (2020) 19:4470–85. doi: 10.1021/acs.jproteome.0c00592
25. Zhu L, Mou C, Yang X, Lin J, Yang Q. Mitophagy in Tgev infection counteracts oxidative stress and apoptosis. Oncotarget. (2016) 7:27122–41. doi: 10.18632/oncotarget.8345
26. Guo L, Yu H, Gu W, Luo X, Li R, Zhang J, et al. Autophagy negatively regulates transmissible gastroenteritis virus replication. Sci Rep. (2016) 6:23864. doi: 10.1038/srep23864
27. Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. (2004) 279:10136–41. doi: 10.1074/jbc.M306124200
28. Duan C, Liu Y, Hao Z, Wang J. Ergosterol peroxide suppresses porcine deltacoronavirus (Pdcov)-induced autophagy to inhibit virus replication via P38 signaling pathway. Vet Microbiol. (2021) 257:109068. doi: 10.1016/j.vetmic.2021.109068
29. Willis S, Day CL, Hinds MG, Huang DC. The Bcl-2-regulated apoptotic pathway. J Cell Sci. (2003) 116(Pt 20):4053–6. doi: 10.1242/jcs.00754
30. Shen Y, Shenk TE. Viruses and apoptosis. Curr Opin Genet Dev. (1995) 5:105–11. doi: 10.1016/S0959-437X(95)90061-6
31. Sun P, Wu H, Huang J, Xu Y, Yang F, Zhang Q, et al. Porcine epidemic diarrhea virus through P53-dependent pathway causes cell cycle arrest in the G0/G1 phase. Virus Res. (2018) 253:1–11. doi: 10.1016/j.virusres.2018.05.019
32. Kim Y, Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. (2014) 460–1:180–93. doi: 10.1016/j.virol.2014.04.040
33. Shen X, Yin L, Pan X, Zhao R, Zhang D. Porcine epidemic diarrhea virus infection blocks cell cycle and induces apoptosis in pig intestinal epithelial cells. Microbial Pathog. (2020) 147:104378. doi: 10.1016/j.micpath.2020.104378
34. Lee C, Kim Y, Jeon JH. Jnk and P38 mitogen-activated protein kinase pathways contribute to porcine epidemic diarrhea virus infection. Virus Res. (2016) 222:1–12. doi: 10.1016/j.virusres.2016.05.018
35. Xu X, Xu Y, Zhang Q, Yang F, Yin Z, Wang L, et al. Porcine epidemic diarrhea virus infections induce apoptosis in vero cells via a reactive oxygen species (Ros)/P53, but Not P38 Mapk and Sapk/Jnk signalling pathways. Vet Microbiol. (2019) 232:1–12. doi: 10.1016/j.vetmic.2019.03.028
36. Ding L, Li J, Li W, Fang Z, Li N, Wu S, et al. P53- and ros-mediated aif pathway involved in Tgev-induced apoptosis. J Vet Med Sci. (2018) 80:1775–81. doi: 10.1292/jvms.18-0104
37. Yang L, Wang C, Shu J, Feng H, He Y, Chen J, et al. Porcine epidemic diarrhea virus induces vero cell apoptosis via the P53-puma signaling pathway. Viruses. (2021) 13:1218. doi: 10.3390/v13071218
38. Chen Y, Zhang Z, Li J, Gao Y, Zhou L, Ge X, et al. Porcine epidemic diarrhea virus S1 protein is the critical inducer of apoptosis. Virol J. (2018) 15:170. doi: 10.1186/s12985-018-1078-4
39. Sun M, Ma J, Yu Z, Pan Z, Lu C, Yao H. Identification of two mutation sites in spike and envelope proteins mediating optimal cellular infection of porcine epidemic diarrhea virus from different pathways. Vet Res. (2017) 48:44. doi: 10.1186/s13567-017-0449-y
40. Si F, Hu X, Wang C, Chen B, Wang R, Dong S, et al. Porcine epidemic diarrhea virus (Pedv) Orf3 enhances viral proliferation by inhibiting apoptosis of infected cells. Viruses. (2020) 12:214. doi: 10.3390/v12020214
41. Kim B, Kim O, Tai JH, Chae C. Transmissible gastroenteritis virus induces apoptosis in swine testicular cell lines but not in intestinal enterocytes. J Comp Pathol. (2000) 123:64–6. doi: 10.1053/jcpa.2000.0386
42. Ding L, Zhao X, Huang Y, Du Q, Dong F, Zhang H, et al. Regulation of Ros in transmissible gastroenteritis virus-activated apoptotic signaling. Biochem Biophys Res Commun. (2013) 442:33–7. doi: 10.1016/j.bbrc.2013.10.164
43. Eleouet JF, Chilmonczyk S, Besnardeau L, Laude H. Transmissible gastroenteritis coronavirus induces programmed cell death in infected cells through a caspase-dependent pathway. J Virol. (1998) 72:4918–24. doi: 10.1128/JVI.72.6.4918-4924.1998
44. Ding L, Xu X, Huang Y, Li Z, Zhang K, Chen G, et al. Transmissible gastroenteritis virus infection induces apoptosis through fasl- and mitochondria-mediated pathways. Vet Microbiol. (2012) 158:12–22. doi: 10.1016/j.vetmic.2012.01.017
45. Huang Y, Ding L, Li Z, Dai M, Zhao X, Li W, et al. Transmissible gastroenteritis virus infection induces cell apoptosis via activation of P53 signalling. J Gen Virol. (2013) 94(Pt 8):1807–17. doi: 10.1099/vir.0.051557-0
46. Eléouët JF, Slee EA, Saurini F, Castagné N, Poncet D, Garrido C, et al. The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (Tgev) is cleaved by caspase-6 and−7 during tgev-induced apoptosis. J Virol. (2000) 74:3975–83. doi: 10.1128/JVI.74.9.3975-3983.2000
47. Ding L, Huang Y, Du Q, Dong F, Zhao X, Zhang W, et al. Tgev nucleocapsid protein induces cell cycle arrest and apoptosis through activation of P53 signaling. Biochem Biophys Res Commun. (2014) 445:497–503. doi: 10.1016/j.bbrc.2014.02.039
48. Jung K, Hu H, Saif LJ. Porcine deltacoronavirus induces apoptosis in swine testicular and Llc porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Vet Microbiol. (2016) 182:57–63. doi: 10.1016/j.vetmic.2015.10.022
49. Lee YJ, Lee C. Porcine deltacoronavirus induces caspase-dependent apoptosis through activation of the cytochrome C-mediated intrinsic mitochondrial pathway. Virus Res. (2018) 253:112–23. doi: 10.1016/j.virusres.2018.06.008
50. Zhang J, Han Y, Shi H, Chen J, Zhang X, Wang X, et al. Swine acute diarrhea syndrome coronavirus-induced apoptosis is caspase- and cyclophilin D- dependent. Emerg Microbes Infect. (2020) 9:439–56. doi: 10.1080/22221751.2020.1722758
51. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. (2006) 124:783–801. doi: 10.1016/j.cell.2006.02.015
52. Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. (2014) 5:461. doi: 10.3389/fimmu.2014.00461
53. Prinz M, Heikenwalder M, Schwarz P, Takeda K, Akira S, Aguzzi A. Prion pathogenesis in the absence of toll-like receptor signalling. EMBO Rep. (2003) 4:195–9. doi: 10.1038/sj.embor.embor731
54. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of Tlr7-mediated recognition of single-stranded RNA. Science. (2004) 303:1529–31. doi: 10.1126/science.1093616
55. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded Rna via toll-like receptor 7 and 8. Science. (2004) 303:1526–9. doi: 10.1126/science.1093620
56. Okahira S, Nishikawa F, Nishikawa S, Akazawa T, Seya T, Matsumoto M. Interferon-beta induction through toll-like receptor 3 depends on double-stranded RNA structure. DNA Cell Biol. (2005) 24:614–23. doi: 10.1089/dna.2005.24.614
57. Hermant P, Michiels T. Interferon-Λ in the context of viral infections: production, response and therapeutic implications. J Innate Immunity. (2014) 6:563–74. doi: 10.1159/000360084
58. Escalante CR, Shen L, Thanos D, Aggarwal AK. Structure of Nf-Kappab P50/P65 heterodimer bound to the Prdii DNA element from the interferon-beta promoter. Structure. (2002) 10:383–91. doi: 10.1016/S0969-2126(02)00723-2
59. Stark GR, Darnell JE Jr. The Jak-Stat pathway at twenty. Immunity. (2012) 36:503–14. doi: 10.1016/j.immuni.2012.03.013
60. Tracz M, Bialek W. Beyond K48 and K63: non-canonical protein ubiquitination. Cell Mol Biol Lett. (2021) 26:1. doi: 10.1186/s11658-020-00245-6
61. Liu Y, Liang QZ, Lu W, Yang YL, Chen R, Huang YW, et al. A comparative analysis of coronavirus nucleocapsid (N) proteins reveals the Sads-Cov N protein antagonizes Ifn-β production by inducing ubiquitination of Rig-I. Front Immunol. (2021) 12:688758. doi: 10.3389/fimmu.2021.688758
62. Likai J, Shasha L, Wenxian Z, Jingjiao M, Jianhe S, Hengan W, et al. Porcine deltacoronavirus nucleocapsid protein suppressed ifn-β production by interfering porcine Rig-I Dsrna-binding and K63-linked polyubiquitination. Front Immunol. (2019) 10:1024. doi: 10.3389/fimmu.2019.01024
63. Ji L, Wang N, Ma J, Cheng Y, Wang H, Sun J, et al. Porcine deltacoronavirus nucleocapsid protein species-specifically suppressed Irf7-induced type I interferon production via ubiquitin-proteasomal degradation pathway. Vet Microbiol. (2020) 250:108853. doi: 10.1016/j.vetmic.2020.108853
64. Sun L, Xing Y, Chen X, Zheng Y, Yang Y, Nichols DB, et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of sting-mediated signaling. PLoS ONE. (2012) 7:e30802. doi: 10.1371/journal.pone.0030802
65. Xing Y, Chen J, Tu J, Zhang B, Chen X, Shi H, et al. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J Gen Virol. (2013) 94(Pt 7):1554–67. doi: 10.1099/vir.0.051169-0
66. Hu X, Tian J, Kang H, Guo D, Liu J, Liu D, et al. Transmissible gastroenteritis virus papain-like protease 1 antagonizes production of interferon-β through its deubiquitinase activity. Biomed Res Int. (2017) 2017:7089091. doi: 10.1155/2017/7089091
67. Wang L, Hu W, Fan C. Structural and biochemical characterization of sads-cov papain-like protease 2. Protein Sci. (2020) 29:1228–41. doi: 10.1002/pro.3857
68. Yin M, Wen W, Wang H, Zhao Q, Zhu H, Chen H, et al. Porcine sapelovirus 3c(Pro) inhibits the production of type I interferon. Front Cell Infect Microbiol. (2022) 12:852473. doi: 10.3389/fcimb.2022.852473
69. Lei X, Han N, Xiao X, Jin Q, He B, Wang J. Enterovirus 71 3c inhibits cytokine expression through cleavage of the Tak1/Tab1/Tab2/Tab3 complex. J Virol. (2014) 88:9830–41. doi: 10.1128/JVI.01425-14
70. Zhang H, Jiang P, Chen Z, Wang D, Zhou Y, Zhu X, et al. Norovirus 3c-like protease antagonizes interferon-β production by cleaving nemo. Virology. (2022) 571:12–20. doi: 10.1016/j.virol.2022.04.004
71. Zhu X, Fang L, Wang D, Yang Y, Chen J, Ye X, et al. Porcine deltacoronavirus Nsp5 inhibits interferon-β production through the cleavage of nemo. Virology. (2017) 502:33–8. doi: 10.1016/j.virol.2016.12.005
72. Wang D, Fang L, Shi Y, Zhang H, Gao L, Peng G, et al. Porcine epidemic diarrhea virus 3c-like protease regulates its interferon antagonism by cleaving nemo. J Virol. (2016) 90:2090–101. doi: 10.1128/JVI.02514-15
73. Zhu X, Wang D, Zhou J, Pan T, Chen J, Yang Y, et al. Porcine deltacoronavirus Nsp5 antagonizes type I interferon signaling by cleaving Stat2. J Virol. (2017) 91(10). doi: 10.1128/JVI.00003-17
74. Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, et al. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between Irf3 and Tbk1. J Virol. (2014) 88:8936–45. doi: 10.1128/JVI.00700-14
75. Chen J, Fang P, Wang M, Peng Q, Ren J, Wang D, et al. Porcine deltacoronavirus nucleocapsid protein antagonizes Ifn-β production by impairing Dsrna and pact binding to Rig-I. Virus Genes. (2019) 55:520–31. doi: 10.1007/s11262-019-01673-z
76. Zhou Z, Sun Y, Xu J, Tang X, Zhou L, Li Q, et al. Swine acute diarrhea syndrome coronavirus nucleocapsid protein antagonizes interferon-β production via blocking the interaction between Traf3 and Tbk1. Front Immunol. (2021) 12:573078. doi: 10.3389/fimmu.2021.573078
77. Zhang J, Yuan S, Peng Q, Ding Z, Hao W, Peng G, et al. Porcine epidemic diarrhea virus Nsp7 inhibits interferon-induced jak-stat signaling through sequestering the interaction between Kpna1 and Stat1. J Virol. (2022) 96:e0040022. doi: 10.1128/jvi.00400-22
78. Gao B, Gong X, Fang S, Weng W, Wang H, Chu H, et al. Inhibition of anti-viral stress granule formation by coronavirus endoribonuclease Nsp15 ensures efficient virus replication. PLoS Pathog. (2021) 17:e1008690. doi: 10.1371/journal.ppat.1008690
79. Yount B, Roberts RS, Sims AC, Deming D, Frieman MB, Sparks J, et al. severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J Virol. (2005) 79:14909–22. doi: 10.1128/JVI.79.23.14909-14922.2005
80. Fang P, Fang L, Ren J, Hong Y, Liu X, Zhao Y, et al. Porcine deltacoronavirus accessory protein Ns6 antagonizes interferon beta production by interfering with the binding of Rig-I/Mda5 to double-stranded Rna. J Virol. (2018) 92:e00712-18. doi: 10.1128/JVI.00712-18
81. Fang P, Fang L, Xia S, Ren J, Zhang J, Bai D, et al. Porcine deltacoronavirus accessory protein Ns7a antagonizes Ifn-β production by competing with Traf3 and Irf3 for binding to Ikkε. Front Cell Infect Microbiol. (2020) 10:257. doi: 10.3389/fcimb.2020.00257
82. Xu X, Zhang H, Zhang Q, Dong J, Liang Y, Huang Y, et al. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol J. (2013) 10:26. doi: 10.1186/1743-422X-10-26
83. Zheng L, Wang X, Guo D, Cao J, Cheng L, Li X, et al. Porcine epidemic diarrhea virus e protein suppresses Rig-I signaling-mediated interferon-β production. Vet Microbiol. (2021) 254:108994. doi: 10.1016/j.vetmic.2021.108994
84. Zheng L, Liu H, Tian Z, Kay M, Wang H, Wang X, et al. Porcine epidemic diarrhea virus E protein inhibits type I interferon production through endoplasmic reticulum stress response (Ers)-mediated suppression of antiviral proteins translation. Res Vet Sci. (2022) 152:236–44. doi: 10.1016/j.rvsc.2022.07.019
85. Shan Y, Liu ZQ, Li GW, Chen C, Luo H, Liu YJ, et al. Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-Λ production by blocking the nuclear factor-κb nuclear translocation. J Zhejiang Univ Sci B. (2018) 19:570–80. doi: 10.1631/jzus.B1700283
86. Li S, Zhu Z, Yang F, Cao W, Yang J, Ma C, et al. Porcine epidemic diarrhea virus membrane protein interacted with Irf7 to inhibit type I Ifn production during viral infection. J Immunol. (2021) 206:2909–23. doi: 10.4049/jimmunol.2001186
87. Wu Z, Cheng L, Xu J, Li P, Li X, Zou D, et al. The accessory protein Orf3 of porcine epidemic diarrhea virus inhibits cellular interleukin-6 and interleukin-8 productions by blocking the nuclear factor-Kb P65 activation. Vet Microbiol. (2020) 251:108892. doi: 10.1016/j.vetmic.2020.108892
88. Zhang Q, Ma J, Yoo D. Inhibition of Nf-Kb Activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology. (2017) 510:111–26. doi: 10.1016/j.virol.2017.07.009
89. Zhang Q, Shi K, Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of Creb-binding protein by Nsp1. Virology. (2016) 489:252–68. doi: 10.1016/j.virol.2015.12.010
90. Zhang Q, Ke H, Blikslager A, Fujita T, Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein Nsp1 in Irf1 signaling. J Virol. (2018) 92:e01677-17. doi: 10.1128/JVI.01677-17
91. Shen Z, Yang Y, Yang S, Zhang G, Xiao S, Fu ZF, et al. Structural and biological basis of alphacoronavirus Nsp1 associated with host proliferation and immune evasion. Viruses. (2020) 12:812. doi: 10.3390/v12080812
92. Wang X, Li H, Li Y, Gao D, Chen L, Chang H, et al. [Effect of porcine epidemic diarrhea virus Nsp1 on type I interferon response]. Sheng Wu Gong Cheng Xue Bao. (2017) 33:1325–34. doi: 10.13345/j.cjb.170068
93. Fan B, Peng Q, Song S, Shi D, Zhang X, Guo W, et al. Nonstructural protein 1 of variant Pedv plays a key role in escaping replication restriction by complement C3. J Virol. (2022) 2022:e0102422. doi: 10.1128/jvi.01024-22
94. Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, et al. Functional screen reveals sars coronavirus nonstructural protein Nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci USA. (2009) 106:3484–9. doi: 10.1073/pnas.0808790106
95. Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, et al. Discovery of an Rna virus 3′->5′ exoribonuclease that is critically involved in coronavirus Rna synthesis. Proc Natl Acad Sci USA. (2006) 103:5108–13. doi: 10.1073/pnas.0508200103
96. Li S, Yang F, Ma C, Cao W, Yang J, Zhao Z, et al. Porcine epidemic diarrhea virus Nsp14 inhibits Nf-Kb pathway activation by targeting the Ikk complex and P65. Anim Dis. (2021) 1:24. doi: 10.1186/s44149-021-00025-5
97. Wang Y, Sun A, Sun Y, Zhang S, Xia T, Guo T, et al. Porcine transmissible gastroenteritis virus inhibits Nf-Kb activity via nonstructural protein 3 to evade host immune system. Virol J. (2019) 16:97. doi: 10.1186/s12985-019-1206-9
98. Liu X, Fang P, Fang L, Hong Y, Zhu X, Wang D, et al. Porcine deltacoronavirus Nsp15 antagonizes interferon-β production independently of its endoribonuclease activity. Mol Immunol. (2019) 114:100–7. doi: 10.1016/j.molimm.2019.07.003
99. Cruz JL, Sola I, Becares M, Alberca B, Plana J, Enjuanes L, et al. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. (2011) 7:e1002090. doi: 10.1371/journal.ppat.1002090
100. Cruz JL, Becares M, Sola I, Oliveros JC, Enjuanes L, Zúñiga S. Alphacoronavirus protein 7 modulates host innate immune response. J Virol. (2013) 87:9754–67. doi: 10.1128/JVI.01032-13
101. Yang L, Xu J, Guo L, Guo T, Zhang L, Feng L, et al. Porcine epidemic diarrhea virus-induced epidermal growth factor receptor activation impairs the antiviral activity of type I interferon. J Virol. (2018) 92:e02095-17. doi: 10.1128/JVI.02095-17
102. Wu Y, Zhang H, Shi Z, Chen J, Li M, Shi H, et al. Porcine epidemic diarrhea virus Nsp15 antagonizes interferon signaling by Rna degradation of Tbk1 and Irf3. Viruses. (2020) 12:599. doi: 10.3390/v12060599
Keywords: SeCoV, apoptosis, autophagy, innate immunity, across species transmission
Citation: Li M, Guo L and Feng L (2022) Interplay between swine enteric coronaviruses and host innate immune. Front. Vet. Sci. 9:1083605. doi: 10.3389/fvets.2022.1083605
Received: 29 October 2022; Accepted: 06 December 2022;
Published: 22 December 2022.
Edited by:
Zhou Mo, Heilongjiang University, ChinaReviewed by:
Huapeng Feng, Zhejiang Sci-Tech University, ChinaXinsheng Liu, Lanzhou Veterinary Research Institute (CAAS), China
Copyright © 2022 Li, Guo and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Longjun Guo,  guolongjun@caas.cn; Li Feng,
guolongjun@caas.cn; Li Feng,  fengli@caas.cn
fengli@caas.cn
 Mingwei Li
Mingwei Li  Longjun Guo
Longjun Guo Li Feng
Li Feng