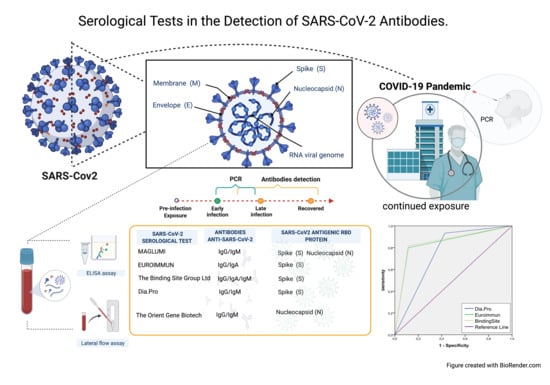
Serological Tests in the Detection of SARS-CoV-2 Antibodies
Abstract
:1. Introduction
2. Methods
2.1. ELISAs
2.2. Lateral Flow Assay (LFA)
2.3. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, Y.-W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.Y.-P.; Lin, R.T.P.; Renia, L.; Ng, L.F.P. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Jin, X. The Progress of 2019 Novel Coronavirus Event in China. J. Med. Virol. 2020, 92, 468–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral Load of SARS-CoV-2 in Clinical Samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- Long, Q.-X.; Liu, B.-Z.; Deng, H.-J.; Wu, G.-C.; Deng, K.; Chen, Y.-K.; Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F.; et al. Antibody Responses to SARS-CoV-2 in Patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, X.; Zhu, C.; Song, Y.; Feng, F.; Qiu, Y.; Feng, J.; Jia, Q.; Song, Q.; Zhu, B.; et al. Immune Phenotyping Based on the Neutrophil-to-Lymphocyte Ratio and IgG Level Predicts Disease Severity and Outcome for Patients with COVID-19. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef]
- Xiao, A.T.; Gao, C.; Zhang, S. Profile of Specific Antibodies to SARS-CoV-2: The First Report. J. Infect. 2020, 81, 147–178. [Google Scholar] [CrossRef]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Asthagiri Arunkumar, G.; Jurczyszak, D.; Polanco, J.; et al. A Serological Assay to Detect SARS-CoV-2 Seroconversion in Humans. medRxiv 2020. [Google Scholar] [CrossRef]
- Estimating SARS-CoV-2 Seroprevalence and Epidemiological Parameters with Uncertainty from Serological Surveys. Available online: https://dash.harvard.edu/handle/1/42659939 (accessed on 17 November 2020).
- Commissioner, O. Coronavirus (COVID-19) Update: FDA Expedites Review of Diagnostic Tests to Combat COVID-19. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-expedites-review-diagnostic-tests-combat-covid-19 (accessed on 17 November 2020).
- Okba, N.M.A.; Müller, M.A.; Li, W.; Wang, C.; Geurtsvan Kessel, C.H.; Corman, V.M.; Lamers, M.M.; Sikkema, R.S.; de Bruin, E.; Chandler, F.D.; et al. SARS-CoV-2 Specific Antibody Responses in COVID-19 Patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Lv, H.; Wu, N.C.; Tsang, O.T.-Y.; Yuan, M.; Perera, R.A.P.M.; Leung, W.S.; So, R.T.Y.; Chan, J.M.C.; Yip, G.K.; Chik, T.S.H.; et al. Cross-Reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020, 31, 107725. [Google Scholar] [CrossRef]
- Carpenter, C.R. Rapid Antigen and Molecular Tests Had Varied Sensitivity and ≥97% Specificity for Detecting SARS-CoV-2 Infection. Ann. Intern. Med. 2020, 173, JC69. [Google Scholar] [CrossRef]
- Ndwandwe, D.; Mathebula, L.; Kamadjeu, R.; Wiysonge, C.S. Cochrane Corner: Rapid Point-of-Care Antigen and Molecular-Based Tests for the Diagnosis of COVID-19 Infection. Pan Afr. Med. J. 2020, 37. [Google Scholar] [CrossRef]
- Deeks, J.J.; Dinnes, J.; Takwoingi, Y.; Davenport, C.; Spijker, R.; Taylor-Phillips, S.; Adriano, A.; Beese, S.; Dretzke, J.; Ferrante di Ruffano, L.; et al. Antibody Tests for Identification of Current and Past Infection with SARS-CoV-2. Cochrane Database Syst. Rev. 2020, 6, CD013652. [Google Scholar] [CrossRef]
- Lin, Y.; Shen, X.; Yang, R.F.; Li, Y.X.; Ji, Y.Y.; He, Y.Y.; Shi, M.D.; Lu, W.; Shi, T.L.; Wang, J.; et al. Identification of an Epitope of SARS-Coronavirus Nucleocapsid Protein. Cell Res. 2003, 13, 141–145. [Google Scholar] [CrossRef]
- Chan, C.M.; Tse, H.; Wong, S.S.Y.; Woo, P.C.Y.; Lau, S.K.P.; Chen, L.; Zheng, B.J.; Huang, J.D.; Yuen, K.Y. Examination of Seroprevalence of Coronavirus HKU1 Infection with S Protein-Based ELISA and Neutralization Assay against Viral Spike Pseudotyped Virus. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2009, 45, 54–60. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and Evolution of Pathogenic Coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet Lond. Eng. 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Miller, T.E.; Garcia Beltran, W.F.; Bard, A.Z.; Gogakos, T.; Anahtar, M.N.; Astudillo, M.G.; Yang, D.; Thierauf, J.; Fisch, A.S.; Mahowald, G.K.; et al. Clinical Sensitivity and Interpretation of PCR and Serological COVID-19 Diagnostics for Patients Presenting to the Hospital. FASEB J. 2020. [Google Scholar] [CrossRef]
- Lisboa Bastos, M.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.-P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic Accuracy of Serological Tests for Covid-19: Systematic Review and Meta-Analysis. BMJ 2020, 370. [Google Scholar] [CrossRef]
- Zainol Rashid, Z.; Othman, S.N.; Abdul Samat, M.N.; Ali, U.K.; Wong, K.K. Diagnostic Performance of COVID-19 Serology Assays. Malays. J. Pathol. 2020, 42, 13–21. [Google Scholar] [PubMed]
- SEI–INMUNOLOGIA–COVID-19. Available online: https://www.inmunologia.org/index.php/covid-19 (accessed on 30 March 2021).
- Ghaffari, A.; Meurant, R.; Ardakani, A. COVID-19 Serological Tests: How Well Do They Actually Perform? Diagn. Basel Switz. 2020, 10, 453. [Google Scholar] [CrossRef] [PubMed]
- Burbelo, P.D.; Riedo, F.X.; Morishima, C.; Rawlings, S.; Smith, D.; Das, S.; Strich, J.R.; Chertow, D.S.; Davey, R.T.; Cohen, J.I. Sensitivity in Detection of Antibodies to Nucleocapsid and Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus 2 in Patients With Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Liu, G.; Ma, H.; Zhao, D.; Yang, Y.; Liu, M.; Mohammed, A.; Zhao, C.; Yang, Y.; Xie, J.; et al. Biochemical Characterization of SARS-CoV-2 Nucleocapsid Protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623. [Google Scholar] [CrossRef]
- Lawandi, A.; Danner, R.L. Antibody Tests Have Higher Sensitivity at ≥15 Days after Symptom Onset and 99% Specificity for Detecting SARS-CoV-2. Ann. Intern. Med. 2020, 173, JC57. [Google Scholar] [CrossRef]
- Nicol, T.; Lefeuvre, C.; Serri, O.; Pivert, A.; Joubaud, F.; Dubée, V.; Kouatchet, A.; Ducancelle, A.; Lunel-Fabiani, F.; Le Guillou-Guillemette, H. Assessment of SARS-CoV-2 Serological Tests for the Diagnosis of COVID-19 through the Evaluation of Three Immunoassays: Two Automated Immunoassays (Euroimmun and Abbott) and One Rapid Lateral Flow Immunoassay (NG Biotech). J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2020, 129, 104511. [Google Scholar] [CrossRef]
- Serrano, M.M.; Rodríguez, D.N.; Palop, N.T.; Arenas, R.O.; Córdoba, M.M.; Mochón, M.D.O.; Cardona, C.G. Comparison of Commercial Lateral Flow Immunoassays and ELISA for SARS-CoV-2 Antibody Detection. J. Clin. Virol. 2020, 129, 104529. [Google Scholar] [CrossRef]
- Lassaunière, R.; Frische, A.; Harboe, Z.B.; Nielsen, A.C.; Fomsgaard, A.; Krogfelt, K.A.; Jørgensen, C.S. Evaluation of Nine Commercial SARS-CoV-2 Immunoassays. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Byrnes, S.A.; Gallagher, R.; Steadman, A.; Bennett, C.; Rivera, R.; Ortega, C.; Motley, S.T.; Jain, P.; Weigl, B.H.; Connelly, J.T. Multiplexed and Extraction-Free Amplification for Simplified SARS-CoV-2 RT-PCR Tests. Anal. Chem. 2021, 93, 4160–4165. [Google Scholar] [CrossRef]
- Liu, Y.; Eggo, R.M.; Kucharski, A.J. Secondary Attack Rate and Superspreading Events for SARS-CoV-2. Lancet Lond. Engl. 2020, 395, e47. [Google Scholar] [CrossRef] [Green Version]




| Test | Anti-SARS-CoV-2 Antibodies Isotype | Serological Assays | Antigenic Region of the RBD Protein |
|---|---|---|---|
| MAGLUMI | IgG/IgM | ELISA | N and S |
| EUROIMMUN | IgG/IgA | ELISA | S (S1) |
| Dia.Pro | IgG/IgM | ELISA | N and S |
| The Binding Site Group Ltd. | IgG/IgA/IgM | ELISA | S (Trimer) |
| The Orient Gene Biotech | IgG/IgM | LFA | N |
| Tests | Samples | Positive (%) | Negative (%) | Total (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Concordance | ||||||||
| Dia.Pro | Binding-Site | Euroimmun | Maglumi | OrientGene | 45 | 55.55 | 4.44 | 60.00 |
| Dia.Pro | Binding-Site | Euroimmun | 132 | 38.63 | 33.33 | 71.97 | ||
| Dia.Pro | Binding-Site | 171 | 36.84 | 38.59 | 75.44 | |||
| Dia.Pro | Euroimmun | 132 | 42.42 | 33.33 | 75.76 | |||
| Binding-Site | Euroimmun | 220 | 29.54 | 15.00 | 44.55 | |||
| Dia.Pro | Maglumi | 45 | 66.66 | 4.44 | 71.11 | |||
| Dia.Pro | OrientGene | 45 | 77.77 | 4.44 | 82.22 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guevara-Hoyer, K.; Fuentes-Antrás, J.; De la Fuente-Muñoz, E.; Rodríguez de la Peña, A.; Viñuela, M.; Cabello-Clotet, N.; Estrada, V.; Culebras, E.; Delgado-Iribarren, A.; Martínez-Novillo, M.; et al. Serological Tests in the Detection of SARS-CoV-2 Antibodies. Diagnostics 2021, 11, 678. https://doi.org/10.3390/diagnostics11040678
Guevara-Hoyer K, Fuentes-Antrás J, De la Fuente-Muñoz E, Rodríguez de la Peña A, Viñuela M, Cabello-Clotet N, Estrada V, Culebras E, Delgado-Iribarren A, Martínez-Novillo M, et al. Serological Tests in the Detection of SARS-CoV-2 Antibodies. Diagnostics. 2021; 11(4):678. https://doi.org/10.3390/diagnostics11040678
Chicago/Turabian StyleGuevara-Hoyer, Kissy, Jesús Fuentes-Antrás, Eduardo De la Fuente-Muñoz, Antonia Rodríguez de la Peña, Marcos Viñuela, Noemí Cabello-Clotet, Vicente Estrada, Esther Culebras, Alberto Delgado-Iribarren, Mercedes Martínez-Novillo, and et al. 2021. "Serological Tests in the Detection of SARS-CoV-2 Antibodies" Diagnostics 11, no. 4: 678. https://doi.org/10.3390/diagnostics11040678