Abstract
Objective
The aims of this study were to develop a multiparametric prognostic model for death in COVID-19 patients and to assess the incremental value of CT disease extension over clinical parameters.
Methods
Consecutive patients who presented to all five of the emergency rooms of the Reggio Emilia province between February 27 and March 23, 2020, for suspected COVID-19, underwent chest CT, and had a positive swab within 10 days were included in this retrospective study. Age, sex, comorbidities, days from symptom onset, and laboratory data were retrieved from institutional information systems. CT disease extension was visually graded as < 20%, 20–39%, 40–59%, or ≥ 60%. The association between clinical and CT variables with death was estimated with univariable and multivariable Cox proportional hazards models; model performance was assessed using k-fold cross-validation for the area under the ROC curve (cvAUC).
Results
Of the 866 included patients (median age 59.8, women 39.2%), 93 (10.74%) died. Clinical variables significantly associated with death in multivariable model were age, male sex, HDL cholesterol, dementia, heart failure, vascular diseases, time from symptom onset, neutrophils, LDH, and oxygen saturation level. CT disease extension was also independently associated with death (HR = 7.56, 95% CI = 3.49; 16.38 for ≥ 60% extension). cvAUCs were 0.927 (bootstrap bias-corrected 95% CI = 0.899–0.947) for the clinical model and 0.936 (bootstrap bias-corrected 95% CI = 0.912–0.953) when adding CT extension.
Conclusions
A prognostic model based on clinical variables is highly accurate in predicting death in COVID-19 patients. Adding CT disease extension to the model scarcely improves its accuracy.
Key Points
• Early identification of COVID-19 patients at higher risk of disease progression and death is crucial; the role of CT scan in defining prognosis is unclear.
• A clinical model based on age, sex, comorbidities, days from symptom onset, and laboratory results was highly accurate in predicting death in COVID-19 patients presenting to the emergency room.
• Disease extension assessed with CT was independently associated with death when added to the model but did not produce a valuable increase in accuracy.
Similar content being viewed by others
Introduction
The clinical course of coronavirus disease 2019 (COVID-19) is variable; most patients experience a mild disease course, but a minority rapidly deteriorates to severe or critical illness [1]. The case fatality rate during the first peak of the pandemic was over 10% in most European countries [2], reaching 20% in our region in the first weeks of the pandemic once 30-day follow-up had been completed for all cases [3, 4]. These rates largely overestimate the real fatality rate of the whole spectrum of SARS-CoV-2 infections because only severe cases were tested by polymerase chain reaction (RT-PCR) in that period. Nevertheless, in March–April 2020 in northern Italy, 30-day survival of hospitalized COVID-19 patients was just over 70% [5].
Early identification of patients at higher risk of disease progression and death is crucial. Effective predictive models based on risk factors for death would help clinicians decide who needs hospitalization and what intensity of care each patient needs. Over the long term, this positive effect on clinical decision-making may improve patient management and outcome and may increase the appropriateness of resource utilization.
Different prognostic models to predict mortality, length of hospital stay, and critical illness have been proposed that use several clinical and laboratory variables, including age, male sex, comorbidities, C-reactive protein, lactic dehydrogenase, and lymphocyte count [1, 6,7,8,9]. However, the studies proposing these models often lacked an adequate description of the study population, were generally conducted on hospitalized patients only, and frequently excluded from any analysis those patients who had neither recovered nor died within the time period considered for outcome assessment [6].
Chest imaging, especially by means of computed tomography (CT), was used during the COVID-19 outbreak to rapidly identify patients with COVID-19 pneumonia while waiting for diagnostic RT-PCR confirmation and to assess disease extension at baseline [10, 11]. Some studies have reported on the potential prognostic value of CT findings, especially disease extension assessed by means of visual estimation or software quantification [12,13,14,15].
The few studies that combined clinical, laboratory, and CT findings in predictive prognostic models [16,17,18,19] showed inconsistent results: some suggested a better performance of the model when adding CT [16]; others showed that CT findings had insufficient prognostic power to be selected in multivariable models [18].
The aims of this study were to develop a multiparametric prognostic model for death in COVID-19 patients and to assess the incremental value of CT disease extension over clinical parameters.
Methods
Setting
In Reggio Emilia province (Northern Italy, 532,000 inhabitants), public hospital care is provided by six hospitals, with five emergency rooms and one radiology department with centralized imaging reading. Access to the emergency room is possible only in public hospitals. The first case of SARS-CoV-2 infection was diagnosed on February 27, 2020. Up to March 24, 2020, there were 1399 RT-PCR-confirmed COVID-19 cases and the daily number of new cases was still rising.
Study design and population
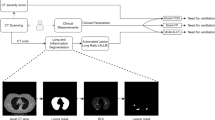
This observational study was approved by the Area Vasta Emilia Nord Ethics Committee on April 7, 2020 (protocol number 2020/0045199). Given the retrospective nature of the study, the Ethics Committee authorizes the use of a patient’s data without his/her informed consent if all reasonable efforts have been made to contact that patient. We included all consecutive patients who presented to the provincial emergency rooms (ERs) between February 27 and March 23, 2020, for suspected COVID-19, underwent chest CT at ER presentation, and were positive on RT-PCR for SARS-CoV-2 within 10 days from ER presentation. Given the organization of emergency hospital assistance in our province and the inclusion criteria, the study design was population-based.
During the COVID-19 outbreak, the diagnostic protocol for suspected COVID-19 patients presenting to the ER included nasopharyngeal/oropharyngeal swabs for RT-PCR, blood tests, chest X-rays, and CT in cases of suggestive X-rays or negative X-rays but with highly suggestive clinical features. A structured CT report was introduced on March 13, 2020. Baseline cross-sectional data of patients presenting to the ER between March 13 and 23, 2020, were used for the assessment of CT diagnostic accuracy in another study [20] which also included patients with negative RT-PCR.
Data collection
Data including date of symptom onset, diagnosis, hospitalization, and death were retrieved from the COVID-19 Surveillance Registry, coordinated by the National Institute of Health and implemented in each Local Health Authority. Registry data were linked with the hospital radiology information system to search for CTs performed at or after the onset of COVID symptoms and with hospital discharge databases to collect information on comorbidities. The Charlson Comorbidity Index (CCI) was used to evaluate the impact of comorbidities. The CCI was calculated based on hospital admissions in the previous 10 years as a weighted measure incorporating 17 different comorbidities, each of which weighted according to its potential impact on mortality [21]. Vascular diseases included atherosclerosis, aortic aneurysms, peripheral artery disease, and peripheral or abdominal embolism. Diabetes was ascertained through linkage with the local Diabetes Registry [22]. Since body mass index (BMI) or other measures of body composition, which are known negative prognostic factors for COVID-19 [23], were not available, the most recent lipid profile (measured in 2015–2018 and including total, low-density lipoprotein (LDL), and high-density lipoprotein (HDL) cholesterol and triglycerides) was used to estimate metabolic risk factors as a pre-existing condition.
Symptoms (fever, cough, dyspnea/polypnea, headache, myalgia/arthralgia, fatigue, syncope, gastrointestinal symptoms, other symptoms) and body temperature at ER presentation were manually collected from medical records only for the subset of patients presenting between February 27 and March 13, 2020. Fever was defined as a body temperature > 37.5 °C, or, when this value was not available, self-reported fever in the preceding days. Dyspnea/polypnea was defined as a respiratory frequency > 18 breaths per minute or reported dyspnea in the preceding 10 days.
Blood tests and RT-PCR
Blood tests at ER presentation were retrieved from the laboratory information system as indicators of clinical condition at admission. The levels of C-reactive protein (CRP), lactate dehydrogenase (LDH), white blood cell, lymphocyte, neutrophil, and platelet counts measured at ER presentation were collected. Oxygen saturation level (SO2) was also collected for patients who had an arterial blood gas analysis before being provided with oxygen support. The tests were carried out in the Hospital Clinical Laboratories with routine automated methods.
To diagnose SARS-CoV-2 infection, a commercial One-Step Reverse Transcription RT-PCR (GeneFinder™ COVID-19 PLUS RealAmp Kit) was used and RT-PCR assay was performed on an Applied Biosystems 7500 Sequence Detection System.
CT acquisition technique
CT scans were performed using one of three different types of scanner present in the five hospitals with an ER (128-slice Somatom Definition Edge, Siemens Healthineers; 64-slice Ingenuity, Philips Healthcare; 16-slice GE BrightSpeed, GE Healthcare) without contrast media injection, with the patient in supine position, during end-inspiration. Scanning parameters were tube voltage 120 kV, automatic tube current modulation, collimation width 0.625 or 1.25 mm, acquisition slice thickness 2.5 mm, and interval 1.25 mm. Images were reconstructed with a high-resolution algorithm at slice thickness 1.0/1.25 mm.
CT structured reporting and retrospective analysis
Between March 13 and March 23, 2020, each radiologist completed a routine CT report and a structured report that included the presence/absence of ground-glass opacities and consolidations, and the extension of pulmonary lesions using a visual scoring system (< 20%, 20–39%, 40–59%, ≥ 60%) [20].
CTs performed between February 27 and March 13, 2020 (before the introduction of the structured report) were retrospectively reviewed by a chest radiologist with 15 years’ experience (L.S.) to collect the same parameters described above, including visual scoring. One hundred consecutive CTs were also reviewed by a radiologist with 3 years’ experience (G.B.) to assess interrater agreement.
Statistical analyses
The sample size was determined by considering the time period in which CT was included in the standard protocol for COVID-19 diagnosis in all suspected patients. Continuous variables were reported as median and interquartile range, and categorical variables as proportions. Single imputation procedure using truncated regression adjusted for sex, age, and comorbidities was used to deal with the problem of any missing values, and missing values were replaced by the mean value of the imputations.
Cox proportional hazards models were used to estimate hazard ratios (HR) with 95% confidence intervals (95% CI) for death, first by univariable model adjusted for age, sex, and calendar time (weeks since the beginning of the outbreak). Statically significant clinical, laboratory, and imaging variables were then used to develop two prognostic multivariable models, with and without CT disease extension. Sensitivity analyses were performed by stratifying the models by time since symptom onset (< 8 and ≥ 8 days) and SO2 levels (< 95% and ≥ 95%) and by excluding patients aged over 85 years, with CT disease extension ≥ 60%, or who had died within 48 h from admission. The performance of the models was assessed using receiver operating characteristic (ROC) curves for 30-day death rate. On the same original sample, we fitted a logit model and used k-fold cross-validation to obtain a bias-corrected estimate of predictive accuracy. This technique averages the AUCs corresponding to each fold and applies the bootstrap procedure to the cross-validated AUC. We used 10 folds, with the exception of sensitivity analyses for ≥ 8 days from symptom onset and SO2 level ≥ 95%, in which we applied 7 and 5 folds respectively, to ensure a minimum outcome frequency in each test set. We reported cross-validated mean AUC (cvAUC), and bootstrap bias-corrected (BBC) 95% CI, with and without CT extension.
We used the kappa-statistic measure of weighted interrater agreement for a double reading of CT disease extension.
We used Stata 13.0 SE (Stata Corporation) software package.
Results
Study population
We included 866 consecutive subjects (median age 59.8, women 39.2%) (Fig. 1), 318 of whom enrolled between February 27 and March 13, 2020. For most of the included patients (809/866), there was a delay between CT scan and RT-PCR of 0 to 2 days (Supplementary Table 1). Median follow-up duration for death was 43 days. During follow-up, 93 (10.74%) patients died (89 in hospital and 4 after discharge to an elderly care residential facility), while 363 (41.92%) were hospitalized.
Patients’ pre-existing conditions, along with clinical, laboratory, and CT findings at ER presentation in the whole cohort and in patients who died and who were hospitalized, are reported in Table 1. Supplementary Table 2 reports the same variables using original data without multiple imputation to address missing values. For CT disease extension, the interrater concordance was excellent (weighted kappa 0.91) (Supplementary Table 3).
Associations between clinical, laboratory, and CT variables with death and hospitalization
Increasing age and male sex were significantly associated with death (HR = 1.11, 95% CI = 1.1–1.1 and HR = 2.6, 95% CI = 1.55–4.36, respectively) and hospitalization (Table 2). Probability of death and hospitalization progressively decreased as the epidemic went on: ER presentation in the first compared with the fourth week of the epidemic had an HR for death of 3.17 (95% CI = 1.39–7.24).
After correcting for age, sex, and calendar time, the Charlson Comorbidity Index was associated with both death and hospitalization (HR = 1.38, 95% CI = 1.17–1.63 and HR = 1.30, 95% CI = 1.17–1.44). Comorbidities significantly associated with death were ischemic cardiopathy, dementia, heart failure, obesity, arrhythmias, and vascular diseases, while diabetes, cancer, COPD, heart failure, arrhythmias, and dementia were associated with hospitalization. Among lipid profile data, low HDL and high triglyceride increase were significantly associated with higher probability of death and of hospitalization.
Time from symptom onset was inversely associated with death (HR = 0.88, 95% CI = 0.83–0.94) and hospitalization. Dyspnea was significantly associated with both death (HR = 1.96, 95% CI = 1.15–3.32) and hospitalization. Among laboratory data, high neutrophil count, CRP and LDH level, and low SO2 were significantly associated with both death and hospitalization.
Presence of consolidation at CT was significantly associated with death (HR = 1.63, 95% CI = 1.00–2.64) and hospitalization (HR = 1.52, 95% CI = 1.20–1.92). High CT visual extension (Fig. 2) was significantly associated with hospitalization for all CT classes and with death for ≥ 60% extension (HR = 6.68, 95% CI = 3.56–12.56).
Multivariable models for death
Variables which were associated with death in the univariable analysis and suitable for a prognostic model to be used in all clinical settings were selected for the multivariable analysis (Table 3); we did not include the variable “calendar period” because its predictive value could not be generalized to other contexts. Demographic and clinical variables that remained significantly associated with death in the multivariable model were age, male sex, HDL cholesterol, dementia, heart failure, vascular diseases, time from symptom onset, neutrophil count, LDH, and SO2. When adding CT disease extension to the clinical model, 40–59% and ≥ 60% extensions were significantly associated with death (HR = 2.41, 95% CI = 1.18–4.93 and HR = 7.56, 95% CI = 3.49–16.38, respectively), while male sex, SO2, and LDH were not.
When evaluating the performance of the two multivariate models in the prediction of death, AUCs were 0.936 (95% CI = 0.916–0.956) and 0.947 (95% CI = 0.930–0.964) respectively for the clinical model and the model including CT extension. By applying cross-validation, the clinical model and the model including CT extension had cvAUCs of 0.927 (BBC 95% CI = 0.899–0.947) and 0.936 (BBC 95% CI = 0.912–0.953) (Fig. 3).
Since previous HDL cholesterol is not always available in daily clinical practice at ER admission, we also built alternative models excluding this variable, obtaining similar cvAUCs (Supplementary Table 4).
Sensitivity analyses and stratifications
When stratifying patients according to time from symptom onset (< or ≥ 8 days) and to SO2 level (SO2 < 95% or ≥ 95%), the cvAUC of the model with CT disease extension remained only slightly higher than the cvAUC of the clinical model (Supplementary Fig. 1; Table 4). In each restricted population, when excluding patients with CT extension ≥ 60%, or excluding patients died within 48 h, or excluding patients older than 85 years, the cvAUC of the model including CT was slightly higher, with the highest increase when excluding patients older than 85 years (cvAUC from 0.929 to 0.943) (Supplementary Fig. 2; Table 4). Both models presented a higher cvAUC in patients with SO2 > 95% (cvAUC 0.952; BBC 95% CI = 0.885–0.966 for the model including CT extension). Finally, when applying the multivariable models to the subcohort of patients with available data on symptoms, cough and gastrointestinal symptoms were significantly associated with death, both in the univariable (Supplementary Table 5) and in the multivariable models without CT disease extension (Supplementary Table 6). When adding CT disease extension, the associations remained similar (HR = 0.56; 95% CI = 0.32–0.99 for cough and HR = 2.70; 95% CI = 1.20–6.10 for gastrointestinal symptoms).
Discussion
A clinical multivariable model based on age, sex, comorbidities, days from symptom onset, and laboratory test results was highly accurate in predicting death in COVID-19 patients presenting to the ER (cvAUC = 0.927). The extension of lung changes evaluated with CT (40–60% and ≥ 60%) was independently associated with death when added to the multivariable model. However, when compared to the clinical model, the model including CT disease extension presented only a slight increase in performance for the prediction of death (cvAUC = 0.936). The slightly added prognostic value was substantially similar when stratifying patients according to time from symptom onset and SO2 level, or when excluding patients with CT extension ≥ 60%, or excluding patients died within 48 h, or excluding patients older than 85 years. Both models performed better in less severe patients in terms of oxygen saturation level (SO2 ≥ 95%).
Some clinical prognostic models have been proposed to predict severe/critical illness: only some had mortality as an outcome, and most were developed and tested on hospitalized patients only. The cvAUC achieved by our clinical model is similar or higher than those previously reported [1, 6, 9, 18].
As in most previous studies [6, 24], age and male sex were strong independent risk factors for death in COVID-19 patients, as was a shorter time between symptom onset and ER presentation. Most considered comorbidities were associated with death, but dementia, heart failure, and peripheral vascular diseases were included in our multivariable model. This result confirms the negative impact of cardiovascular comorbidities on COVID-19 prognosis [24, 25], while dementia is probably an indicator of frailty and poor nutritional status, which are typical of older patients, many of whom live in nursing homes. Since the role of metabolic derangement in COVID-19 patients has been underlined [25, 26], we evaluated previous lipid profile. Both high triglycerides and low HDL cholesterol levels were associated with mortality, the latter being included in the predictive model. Among laboratory tests, variables included in the model were neutrophil count, LDH, and SO2, which reflect different disease pathways and manifestations, and confirming the results of previous studies [6, 9, 16].
The potential prognostic role of CT-based assessment of lung disease extension has been suggested [12,13,14,15, 27], and a few studies have included it in combined prognostic models [16,17,18,19]. Colombi et al found a slightly higher increase in AUC (from 0.83 to 0.86) when adding CT disease extension to a clinical model predictive of intensive care unit admission and/or death [16]. The main differences with our study are the outcome, the use of a logistic regression analysis rather than a Cox proportional hazards model, and a different CT assessment method, which focused on well-aerated lung. Finally, we used the cross-validation method to account for the lack of an external validation set. Interestingly, the clinical model proposed by Colombi et al, unlike ours, did not include SO2.
In our study, when adding CT disease extension to the clinical model, SO2 and LDH were no longer significantly associated with mortality. This seems to point out that CT disease extension and SO2 may describe the same phenomenon, with CT allowing a small gain in informational value, possibly because CT lung changes precede a clinically significant SO2 decrease. However, our results, particularly the high AUC of the clinical model and the limited improvement in its performance when adding CT disease extension, do not seem to support the routine use of CT for prognostic purposes in COVID-19 patients. Nevertheless, our results are limited to short-term outcomes, while the role of CT for the evaluation of long-term outcomes needs to be more fully explored [28].
The major limitation of this study is the lack of a validation cohort to test the generalizability of the model. Nevertheless, in the statistical analyses we adopted the cross-validation, an internal validation approach for prediction models, used to generate a more realistic estimate of predictive performance when the number of observations is not very large [29]. Moreover, although we propose a prognostic model to be applied to COVID-19 patients presenting to the ER, the study population was somewhat selected: a small percentage of patients presenting to the ER did not undergo a CT. This selection may have introduced a bias if the decision to perform a CT had been influenced by factors, including comorbidities, which we used in the prognostic model. If this selection occurred, the consequence is an underestimation of the prognostic value of comorbidities associated with both the probability of being referred to CT and with death. However, since hospitalization is surely more influenced by these factors than the decision to perform a CT scan, this kind of bias affects more the studies conducted on hospitalized patients only. Due to the retrospective nature of the study, some data were not easily available: arterial blood gas analysis rather than pulse oximeter results was used to obtain oxygen saturation level, and symptoms were collected for a subset of patients only. We used the most recent lipid profile to evaluate patients’ metabolic conditions, but we lacked BMI, which is generally easier to obtain in clinical practice. Finally, this study was conducted in an early phase of the epidemic, before that the prognostic role of other laboratory data, above all D-dimer [30], had emerged. These other tests were therefore not collected in this cohort.
In conclusion, a prognostic model based on sex, age, pre-existing clinical features, days from symptom onset, and laboratory test results at ER presentation is highly accurate in predicting mortality in COVID-19. CT-assessed disease extension is an independent predictive factor for mortality, but the increase in performance of the prognostic model when including CT extension is limited. Further studies are needed to assess the generalizability of the prognostic model.
Abbreviations
- BBC:
-
Bootstrap bias-corrected
- BMI:
-
Body mass index
- CI:
-
Confidence interval
- COPD:
-
Chronic obstructive pulmonary disease
- COVID-19:
-
Coronavirus disease 2019
- CRP:
-
C-reactive protein
- cvAUC:
-
Cross-validated area under the ROC curve
- HDL:
-
High-density lipoprotein
- HR:
-
Hazard ratio
- LDH:
-
Lactate dehydrogenase
- LDL:
-
Low-density lipoprotein
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- SO2:
-
Oxygen saturation level
References
Gong J, Ou J, Qiu X et al (2020) A tool for early prediction of severe coronavirus disease 2019 (COVID-19): a multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin Infect Dis 71(15):833–840. https://doi.org/10.1093/cid/ciaa443
Dong E, Du H, Gardner L (2020) An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20(5):533–534. https://doi.org/10.1016/S1473-3099(20)30120-1 Erratum in: Lancet Infect Dis. 2020 Sep;20(9):e215
Giorgi Rossi P, Broccoli S, Angelini P et al (2020) Case fatality rate in patients with COVID-19 infection and its relationship with length of follow up. J Clin Virol 128:104415. https://doi.org/10.1016/j.jcv.2020.104415
Giorgi Rossi P, Marino M, Formisano D et al (2020) Characteristics and outcomes of a cohort of COVID-19 patients in the Province of Reggio Emilia, Italy. PLoS One 15(8):e0238281. https://doi.org/10.1371/journal.pone.0238281
Ferroni E, Giorgi Rossi P, Spila Alegiani S et al (2020) Survival of hospitalized COVID-19 patients in Northern Italy: a population-based cohort study by the ITA-COVID-19 Network. Clin Epidemiol 12:1337–1346. https://doi.org/10.2147/CLEP.S271763
Wynants L, Van Calster B, Collins GS et al (2020) Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. BMJ 369:m1328. https://doi.org/10.1136/bmj.m1328 Erratum in: BMJ. 2020 Jun 3;369:m2204
Chen R, Liang W, Jiang M et al (2020) Risk factors of fatal outcome in hospitalized subjects with coronavirus disease 2019 from a nationwide analysis in China. Chest. 158(1):97–105. https://doi.org/10.1016/j.chest.2020.04.010
Ji D, Zhang D, Xu J et al (2020) Prediction for progression risk in patients with COVID-19 pneumonia: the CALL Score. Clin Infect Dis ciaa414. https://doi.org/10.1093/cid/ciaa414
Liang W, Liang H, Ou L et al (2020) Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med 180(8):1–9. https://doi.org/10.1001/jamainternmed.2020.2033
Rubin GD, Ryerson CJ, Haramati LB et al (2020) The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Chest. 158(1):106–116. https://doi.org/10.1016/j.chest.2020.04.003
Revel MP, Parkar AP, Prosch H et al (2020) COVID-19 patients and the radiology department - advice from the European Society of Radiology (ESR) and the European Society of Thoracic Imaging (ESTI). Eur Radiol 30(9):4903–4909. https://doi.org/10.1007/s00330-020-06865-y
Lyu P, Liu X, Zhang R, Shi L, Gao J (2020) The performance of chest CT in evaluating the clinical severity of COVID-19 pneumonia: identifying critical cases based on CT characteristics. Invest Radiol 55(7):412–421. https://doi.org/10.1097/RLI.0000000000000689
Li Y, Yang Z, Ai T, Wu S, Xia L (2020) Association of “initial CT” findings with mortality in older patients with coronavirus disease 2019 (COVID-19). Eur Radiol:1–8. https://doi.org/10.1007/s00330-020-06969-5 Erratum in: Eur Radiol. 2020 Jul 28
Wang S, Zha Y, Li W et al (2020) A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur Respir J 56(2):2000775. https://doi.org/10.1183/13993003.00775-2020
Yuan M, Yin W, Tao Z, Tan W, Hu Y (2020) Association of radiologic findings with mortality of patients infected with 2019 novel coronavirus in Wuhan, China. PLoS One 15(3):e0230548. https://doi.org/10.1371/journal.pone.0230548
Colombi D, Bodini FC, Petrini M et al (2020) Well-aerated lung on admitting chest CT to predict adverse outcome in COVID-19 pneumonia. Radiology. 296(2):E86–E96. https://doi.org/10.1148/radiol.2020201433
Xu PP, Tian RH, Luo S et al (2020) Risk factors for adverse clinical outcomes with COVID-19 in China: a multicenter, retrospective, observational study. Theranostics. 10(14):6372–6383
Wu G, Yang P, Xie Y et al (2020) Development of a clinical decision support system for severity risk prediction and triage of COVID-19 patients at hospital admission: an international multicentre study. Eur Respir J 56(2):2001104. https://doi.org/10.1183/13993003.01104-2020
Yu T, Cai S, Zheng Z et al (2020) Association Between Clinical Manifestations and Prognosis in Patients with COVID-19. Clin Ther 42(6):964–972. https://doi.org/10.1016/j.clinthera.2020.04.009
Besutti G, Giorgi Rossi P, Iotti V et al (2020) Accuracy of CT in a cohort of symptomatic patients with suspected COVID-19 pneumonia during the outbreak peak in Italy. Eur Radiol 30(12):6818–6827. https://doi.org/10.1007/s00330-020-07050-x
Charlson ME, Pompei P, Ales KL, MacKenzie CR (1987) A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40(5):373–383
Ballotari P, Chiatamone Ranieri S, Vicentini M et al (2014) Building a population-based diabetes register: an Italian experience. Diabetes Res Clin Pract 103(1):79–87. https://doi.org/10.1016/j.diabres.2013.11.020
Simonnet A, Chetboun M, Poissy J et al (2020) High prevalence of obesity in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity (Silver Spring) 28(7):1195–1199. https://doi.org/10.1002/oby.22831 Erratum in: Obesity (Silver Spring). 2020 Oct;28(10):1994
Zheng Z, Peng F, Xu B et al (2020) Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect 81(2):e16–e25. https://doi.org/10.1016/j.jinf.2020.04.021
Li B, Yang J, Zhao F et al (2020) Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin Res Cardiol 109(5):531–538. https://doi.org/10.1007/s00392-020-01626-9
Watanabe M, Risi R, Tuccinardi D, Baquero CJ, Manfrini S, Gnessi L (2020) Obesity and SARS-CoV-2: a population to safeguard. Diabetes Metab Res Rev e3325. https://doi.org/10.1002/dmrr.3325
Lanza E, Muglia R, Bolengo I et al (2020) Quantitative chest CT analysis in COVID-19 to predict the need for oxygenation support and intubation. Eur Radiol 26:1–9. https://doi.org/10.1007/s00330-020-07013-2
Fu Z, Tang N, Chen Y et al (2020) CT features of COVID-19 patients with two consecutive negative RT-PCR tests after treatment. Sci Rep 10(1):11548. https://doi.org/10.1038/s41598-020-68509-x
LeDell E, Petersen M, van der Laan M (2015) Computationally efficient confidence intervals for cross-validated area under the ROC curve estimates. Electron J Stat 9(1):1583–1607. https://doi.org/10.1214/15-EJS1035
Zhang L, Yan X, Fan Q et al (2020) D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J Thromb Haemost 18(6):1324–1329. https://doi.org/10.1111/jth.14859
Acknowledgements
We thank Jacqueline Costa for the English language editing. This study is part of a larger project supported by Ministry of Health (Grant COVID-2020-12371808).
The following are the members of the Working group: Massimo Costantini, Roberto Grilli, Massimiliano Marino, Giulio Formoso, Debora Formisano, Paolo Giorgi Rossi, Emanuela Bedeschi, Cinzia Perilli, Elisabetta La Rosa, Eufemia Bisaccia, Ivano Venturi, Massimo Vicentini, Cinzia Campari, Francesco Gioia, Serena Broccoli, Marta Ottone, Pierpaolo Pattacini, Giulia Besutti, Valentina Iotti, Lucia Spaggiari, Pamela Mancuso, Andrea Nitrosi, Marco Foracchia, Rossana Colla, Alessandro Zerbini, Marco Massari, Anna Maria Ferrari, Mirco Pinotti, Nicola Facciolongo, Ivana Lattuada, Laura Trabucco, Stefano De Pietri, Giorgio Francesco Danelli, Laura Albertazzi, Enrica Bellesia, Simone Canovi, Mattia Corradini, Tommaso Fasano, Elena Magnani, Annalisa Pilia, Alessandra Polese, Silvia Storchi Incerti, Piera Zaldini, Efrem Bonelli, Bonanno Orsola, Matteo Revelli, Carlo Salvarani, Carmine Pinto, Francesco Venturelli.
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Paolo Giorgi Rossi.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
Two of the authors have significant statistical expertise (Marta Ottone, Paolo Giorgi Rossi).
Informed consent
Written informed consent was obtained from all subjects (patients) in the study.
Ethical approval
Institutional Review Board approval was obtained.
Study subjects or cohorts overlap
Baseline cross-sectional data of patients presenting to the ER between March 13 and 23, 2020, were used for the assessment of CT diagnostic accuracy in another study [Besutti G, Eur Radiol 2020] which also included patients with negative RT-PCR.
Besutti G, Giorgi Rossi P, Iotti V, Spaggiari L, Bonacini R, Nitrosi A, Ottone M, Bonelli E, Fasano T, Canovi S, Colla R, Massari M, Lattuada IM, Trabucco L, Pattacini P; Reggio Emilia COVID-19 Working Group. Accuracy of CT in a cohort of symptomatic patients with suspected COVID-19 pneumonia during the outbreak peak in Italy. Eur Radiol. 2020 Jul 14:1–10. doi: 10.1007/s00330-020-07050-x.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 157 kb)
Rights and permissions
About this article
Cite this article
Besutti, G., Ottone, M., Fasano, T. et al. The value of computed tomography in assessing the risk of death in COVID-19 patients presenting to the emergency room. Eur Radiol 31, 9164–9175 (2021). https://doi.org/10.1007/s00330-021-07993-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-07993-9