Published online Oct 6, 2021. doi: 10.12998/wjcc.v9.i28.8388
Peer-review started: April 13, 2021
First decision: May 11, 2021
Revised: May 12, 2021
Accepted: August 16, 2021
Article in press: August 16, 2021
Published online: October 6, 2021
The novel coronavirus disease 2019 (COVID-19) pandemic is a global threat caused by the severe acute respiratory syndrome coronavirus-2.
To develop and validate a risk stratification tool for the early prediction of intensive care unit (ICU) admission among COVID-19 patients at hospital admission.
The training cohort included COVID-19 patients admitted to the Wuhan Third Hospital. We selected 13 of 65 baseline laboratory results to assess ICU admission risk, which were used to develop a risk prediction model with the random forest (RF) algorithm. A nomogram for the logistic regression model was built based on six selected variables. The predicted models were carefully calibrated, and the predictive performance was evaluated and compared with two previously published models.
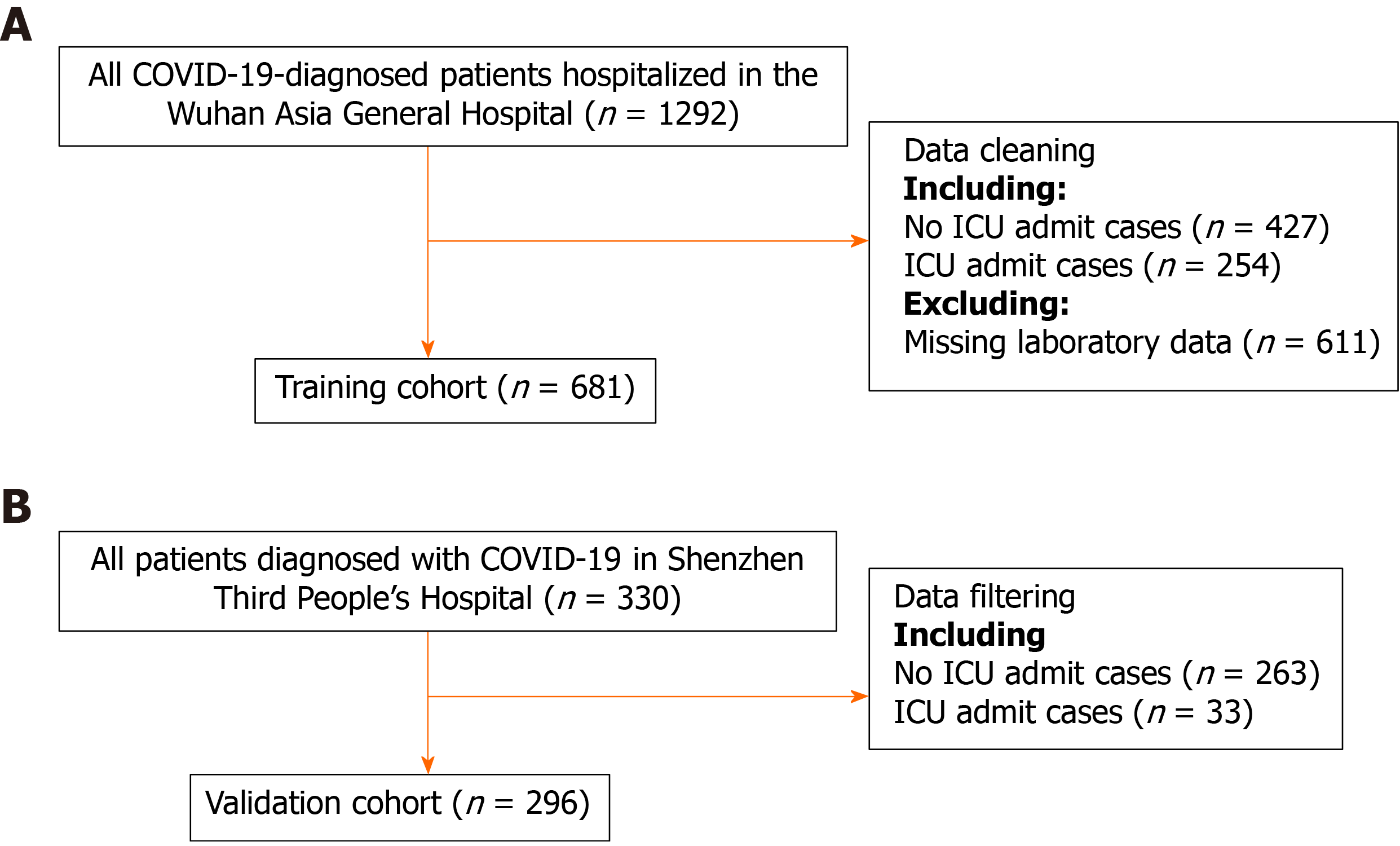
There were 681 and 296 patients in the training and validation cohorts, respectively. The patients in the training cohort were older than those in the validation cohort (median age: 63.0 vs 49.0 years, P < 0.001), and the percentages of male gender were similar (49.6% vs 49.3%, P = 0.958). The top predictors selected in the RF model were neutrophil-to-lymphocyte ratio, age, lactate dehydrogenase, C-reactive protein, creatinine, D-dimer, albumin, procalcitonin, glucose, platelet, total bilirubin, lactate and creatine kinase. The accuracy, sensitivity and specificity for the RF model were 91%, 88% and 93%, respectively, higher than those for the logistic regression model. The area under the receiver operating characteristic curve of our model was much better than those of two other published methods (0.90 vs 0.82 and 0.75). Model A underestimated risk of ICU admission in patients with a predicted risk less than 30%, whereas the RF risk score demonstrated excellent ability to categorize patients into different risk strata. Our predictive model provided a larger standardized net benefit across the major high-risk range compared with model A.
Our model can identify ICU admission risk in COVID-19 patients at admission, who can then receive prompt care, thus improving medical resource allocation.
Core Tip: This study established a risk stratification tool for the early prediction of intensive care unit admission among coronavirus disease 2019 patients at hospital admission to enable such patients to receive immediate appropriate care, thus improving medical resource allocation. The model with 13 indicators selected from 65 laboratory results collected at hospital admission could be used to assess the risk of intensive care unit admission. This study provided a simple probability prediction model to identify intensive care unit admission risk in coronavirus disease 2019 patients at admission.
- Citation: Huang HF, Liu Y, Li JX, Dong H, Gao S, Huang ZY, Fu SZ, Yang LY, Lu HZ, Xia LY, Cao S, Gao Y, Yu XX. Validated tool for early prediction of intensive care unit admission in COVID-19 patients. World J Clin Cases 2021; 9(28): 8388-8403
- URL: https://www.wjgnet.com/2307-8960/full/v9/i28/8388.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i28.8388
The coronavirus disease 2019 (COVID-19) outbreak started in Wuhan, China in December 2019[1,2]. Since then, COVID-19 spread rapidly to pandemic proportions. This disease is caused by severe acute respiratory syndrome coronavirus 2. The disease is associated with symptoms of varying severity. While some patients remain asymptomatic, some exhibit more severe symptoms that rapidly progress to acute respiratory distress syndrome, metabolic acidosis, coagulopathy and septic shock[3,4]. Therefore, patients with severer forms of the disease often require intensive care unit (ICU) care.
The severity and prognosis of COVID-19 varies widely. The clinical characteristics of COVID-19 that impact the disease course can serve as a guide in clinical decision-making[5,6]. Currently, COVID-19 research has focused on the epidemiology and the clinical characteristics of patients[3,7]; however, very few studies have reported the early prediction of prognosis, especially in terms of disease course severity or probability of ICU admission.
Due to the rapidly expanding number of patients and the limited resources in the ICU, prediction models for COVID-19 are crucial in clinical decision-making and medical resource micro-allocation. However, although approximately 50 prognostic models have been built so far, including eight models to predict progression to severe or critical disease[8], only four of the models predicted ICU admission[9-12]. Among the four studies, only two calibrated their models, resulting in underestimation of the risk of poor outcomes and miscalibration risks during external validation[11,12]. Several prognostic predictive models mainly based on laboratory tests have been developed to predict disease progression to a severe or critical state, and the estimated C index of model performance was approximately 0.85[13-15]. Similarly, one of these studies reported perfect calibration. However, the method to check calibration may have been suboptimal[8].
At the start of the pandemic, there was no antiviral agent or vaccine that existed to target this virus, and none of the existing antiretroviral treatments had been recommended for this disease. On October 22, 2020, remdesivir was approved by the Food and Drug Administration as a drug for treating hospitalized COVID-19 patients aged 12 years or more[16]. Since then, ledipasvir and paritaprevir, which have been approved by the Food and Drug Administration, have also been shown to have potential in the treatment of COVID-19. The United States Food and Drug Administration has granted Emergency Use Authorization for the use of two messenger RNA vaccines against COVID-19[17]. However, even highly effective vaccines cannot keep the pandemic under check unless they cover a high percentage of the population. Therefore, it is necessary to stratify patients by illness severity risk or ICU admission risk so that patients who are at higher risk of requiring ICU admission can be identified; this can help reduce the burden of ICU usage, particularly in resource-limited settings. The development of a prognostic model is crucial to address the problem of micro-allocation of scarce healthcare resources in the face of a pandemic[18].
Therefore, the present study aimed to develop and validate a risk stratification tool for the early prediction of ICU admission among COVID-19-positive patients with reference to previously published literature and expert opinion together with data-driven methods. To this end, we externally validated the predicted model on another dataset, and its performance was carefully calibrated.
In this retrospective study, all patients with a positive COVID-19 diagnosis according to any one of the following diagnostic criteria were included in the present study: (1) Respiratory tract or blood specimens were positive for severe acute respiratory syndrome coronavirus 2 nucleic acids by real-time fluorescence reverse transcription-polymerase chain reaction; (2) Genetic sequencing of respiratory tract or blood specimens revealed that the material had high homology with severe acute respiratory syndrome coronavirus 2; and (3) Suspected cases with imaging features of pneumonia consistent with that described in the “Diagnosis and treatment plan for pneumonia infected with new coronavirus [trial version 5]” issued by the National Health Commission of China (this standard was limited to Hubei Province).
Consecutive patients diagnosed with COVID-19 in the Wuhan Third Hospital between December 2019 and March 2020 were included in the training cohort. There were 681 patients in the training cohort. The predictive model was built and internally validated using the above data. The features evaluated for the predictive model included baseline demographics and laboratory data of each patient obtained at their first examination after admission. All blood and urinary samples were processed within 2 h of collection. Figure 1 presents a flowchart illustrating the patients in the training and validation cohorts. The data for each cohort was obtained and analyzed retrospectively. Cases in need of ICU admission were defined according to the following criteria[19]: (1) Respiratory rate ≥ 30 times/min; (2) Pulse oximeter oxygen saturation ≤ 93% at rest; and (3) Partial pressure of arterial oxygen/fraction of inspired oxygen ≤ 300 mmHg.
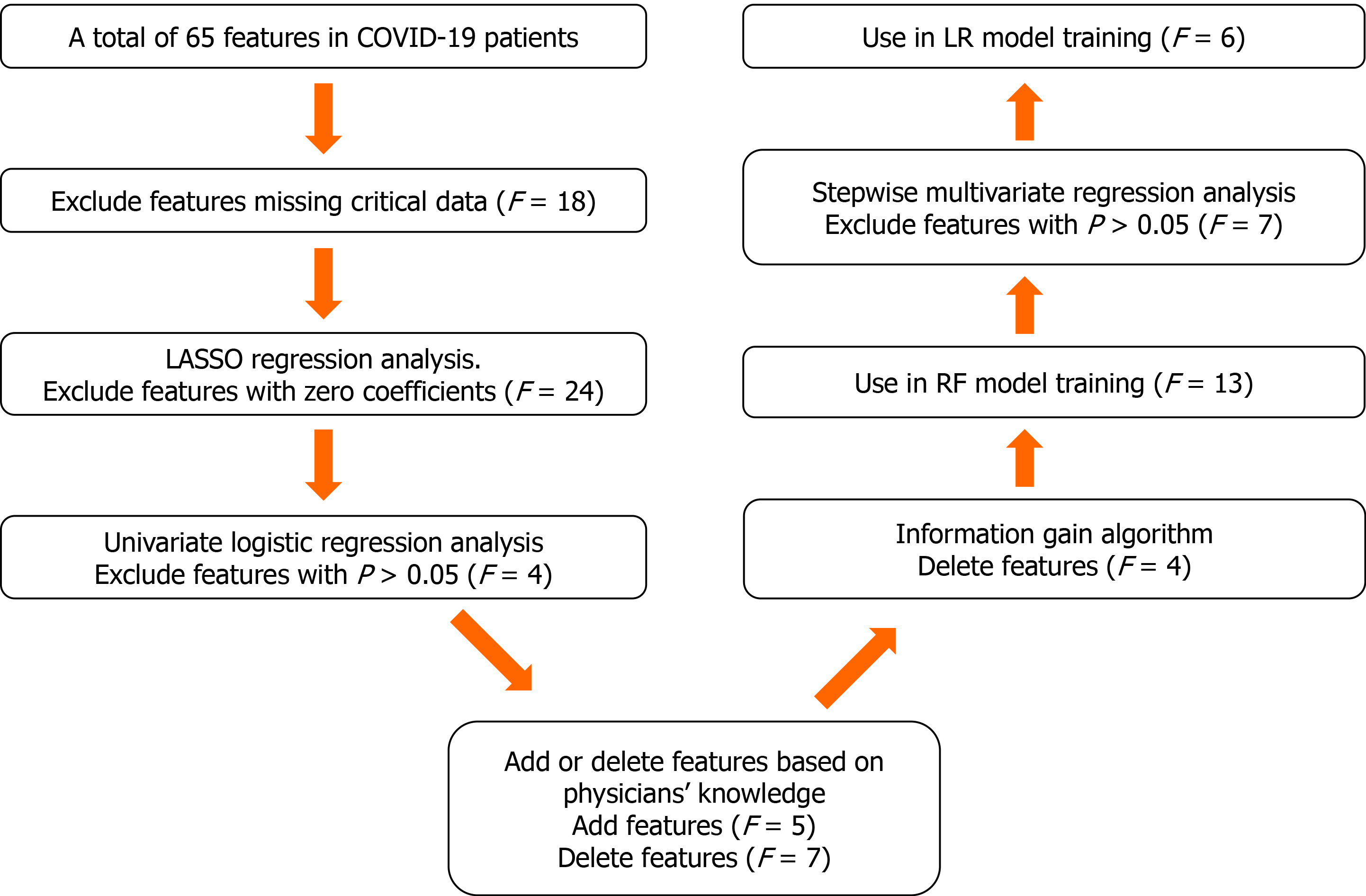
The proposed algorithm used in this study was built on the basis of the random forest (RF) algorithm[20], with modifications made to improve the selection of features (Figure 2). The number of trees was set to 480, and the number of variables selected at each split was set to 4.
Feature selection consisted of the following two steps. First, the least absolute shrinkage and selection operator logistic regression (LR) and univariate LR were used to determine which variables were associated with disease prognosis. We performed a tenfold cross-validation of the training set to calculate the weight of least absolute shrinkage and selection operator penalty. Furthermore, physicians’ knowledge, together with previously published predictors significantly associated with COVID-19 severity[21] were also used to guide feature selection. Then, since each feature’s relative rank could reflect its relative significance[22-24], the information gain algorithm based on entropy and out-of-bag error assessment were used to screen the selected variables for training the RF model. Furthermore, we carried out stepwise multivariate regression analysis to screen the selected variables for training the LR model. We used variables with a P < 0.05 to build the model.
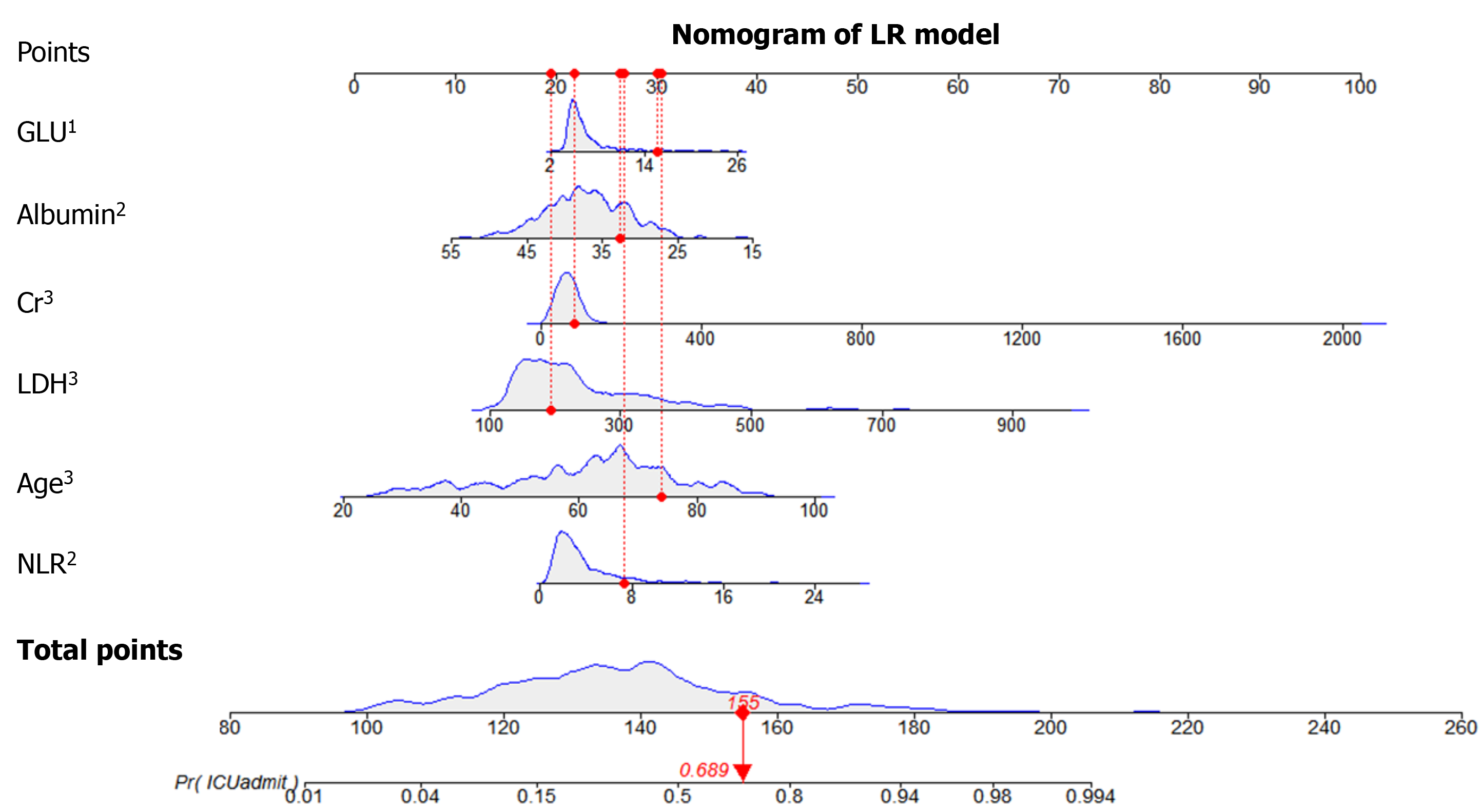
A tree-based ensembled machine learning algorithm, RF, was used to build a risk prediction model based on 13 selected variables. GridsearchCV was performed to search the best parameter for the optimal model. A nomogram for the LR model was built based on six selected variables, which were then screened by multivariate logistic regression analysis. The models were developed in Python version 3.6.5. The reporting followed the TRIPOD statements[25].
Here, we comparatively assessed the predictive performances of scores yielded by the present and conventionally used models, as described below:
Discrimination: To evaluate discrimination, we used the area under the receiver operating characteristic curve, accuracy, specificity, sensitivity, box plots of predicted probabilities of ICU admission and corresponding discrimination slopes, defined as the differences between the mean predicted risks for ICU admission.
Calibration: The conventional Hosmer-Lemeshow statistic was avoided due to its shortcomings[26,27]. Calibration of the predictive model was assessed by the visual representation of the relationship between the predicted and observed values[28]. We constructed calibration curves by plotting the predicted risk of ICU admission divided into 20 groups based on the model risk score against the observed ICU admission.
The net reclassification index (NRI), which has been devised to overcome the limitations of usual discrimination and calibration measures, was computed to compare our proposed algorithm to the other scores[29]. The NRI comparing risk score A of ICU admission to score B was defined as two times the difference between the proportion of no ICU admission and ICU admission groups, respectively, which deemed the risk of ICU admission to be higher according to score A than according to score B[30]. Positive values of the NRI indicated that score A had better discriminative ability than score B, whereas negative values indicated the opposite.
A decision curve analysis (DCA) was used to estimate the clinical usefulness and net benefit of the intervention[31]. The decision curve[32] is a novel and clever graphical device used to assess the potential population impact of adopting a risk prediction instrument into clinical practice. It is grounded in a decision-theoretical framework that accounts for both the benefits of intervention and the costs of intervention to a patient who cannot benefit from the intervention.
A completely independent dataset was then used to externally validate the predictive performance of the algorithm developed herein. To this end, we randomly collected patients with COVID-19 that had been clinically confirmed by reverse transcription-polymerase chain reaction between January 19, 2020 and March 14, 2020, in Shenzhen Third People’s Hospital, which is a tertiary-care teaching hospital. Informed consents were obtained from all patients or from their families by telephone before their data were used in this study. All patient privacy data were protected under the confidentiality policy. Data were analyzed using the statistical software package R, version 3.4.3 (R Core Team, 2017) and EmpowerStats (X&Y solutions, Inc. Boston, Massachusetts).
The performance of the proposed predictive model was compared to other recently published models on the same external validation data. For convenience, the two published models were designated model A[33] and model B[34].
All participants’ baseline demographic and clinical characteristics were obtained at admission. Continuous variables were presented as means ± SD or medians (interquartile ranges), whereas categorical variables were presented using frequencies (percentages). Intergroup differences were analyzed with the χ² test, one-way analysis of variance and Kruskal-Wallis test for categorical variables, normal variables and continuous variables with skewed distribution, respectively. A P < 0.05 was considered statistically significant.
This study describes the development of an algorithm for the early prediction of ICU admission among COVID-19 patients at hospital admission. For developing the prediction model, we first used a training cohort consisting of 681 patients from Wuhan Third Hospital and analyzed their basic baseline demographic and laboratory data obtained at the first admission. We then calibrated the performance of this prediction tool using an entirely different sample set of 296 patients from Shenzhen Third People’s Hospital.
Table 1 presents a comparison of the baseline clinical and laboratory characteristics between the training and validation cohorts. The patients in the training cohort were older than those in the validation cohort (median age: 63.0 vs 49.0 years, P < 0.001), and the percentages of male gender were similar (49.6% vs 49.3%, P = 0.958). There was also some heterogeneity in laboratory results among the different patient groups. Table 1 presents all the patient characteristics.
| Characteristics | Training cohort, n = 681 | Validation cohort, n = 296 | P value |
| Age (yr) | 63.0 (51.0-71.0) | 49.0 (36.0-61.0) | < 0.001 |
| Gender (male) | 338 (49.6%) | 146 (49.3%) | 0.985 |
| BNP (ng/L) | 27.9 (11.5-64.5) | 37.6 (37.6-37.8) | < 0.001 |
| CRP (mg/L) | 13.7 (2.5-53.6) | 11.4 (5.0-26.2) | 0.073 |
| D-dimer (mg/L) | 0.6 (0.3-1.4) | 0.4 (0.3-0.5) | < 0.001 |
| Albumin (g/L) | 37.4 (33.7-40.8) | 43.0 (40.7-44.8) | < 0.001 |
| C3 (g/L) | 1.1 (1.0-1.2) | 1.1 (1.1-1.1) | 0.382 |
| AST (U/L) | 27.0 (20.0-38.0) | 26.3 (21.0-34.2) | 0.230 |
| APTT (s) | 29.7 (26.5-33.5) | 35.1 (32.4-37.8) | < 0.001 |
| K+ (mmol/L) | 3.9 (3.6-4.3) | 3.8 (3.6-4.0) | < 0.001 |
| LYMPH (109/L) | 1.1 (0.8-1.5) | 1.3 (1.0-1.7) | < 0.001 |
| IgA (g/L) | 2.6 (2.0-3.3) | 2.0 (2.0-2.0) | < 0.001 |
| LDH (U/L) | 214.0 (169.0-294.0) | 224.0 (179.0-397.8) | 0.001 |
| Hgb (g/L) | 126.0 (116.0-135.0) | 137.0 (126.0-146.0) | < 0.001 |
| GLU (mmoL/L) | 5.4 (4.7-6.8) | 5.8 (5.3-6.6) | < 0.001 |
| NEUT (109/L) | 3.2 (2.4-4.4) | 2.8 (2.0-3.6) | < 0.001 |
| CK (U/L) | 71.0 (46.0-133.0) | 61.0 (61.0-61.0) | 0.001 |
| Cr (μmoI/L) | 65.9 (53.6-81.5) | 63.0 (52.5-77.0) | 0.033 |
| ALT (U/L) | 24.0 (15.0-37.0) | 24.0 (16.0-35.3) | 0.465 |
| PLT (109/L) | 192.0 (152.0-258.0) | 184.0 (143.8-227.0) | 0.002 |
| PCT (ug/L) | 0.05 (0.05-0.06) | 0.04 (0.03-0.06) | < 0.001 |
| TBil (μmol/L) | 8.9 (6.6-11.9) | 10.2 (7.9-13.9) | < 0.001 |
| WBC (109/L) | 5.0 (3.9-6.3) | 4.7 (3.7-5.8) | 0.032 |
| NLR | 2.8 (1.9-4.5) | 2.1 (1.4-3.2) | < 0.001 |
| LAC (mmoL/L) | 2.8 (2.3-3.4) | 1.4 (1.1-1.5) | < 0.001 |
We selected a total of 65 baseline clinical features for use in our prediction tool. Those with missing values were deleted (n = 18), and the remaining 47 features with complete data were used as potential predictors of critical illness requiring ICU admission. Twenty-three predictors with non-zero coefficients were selected in the least absolute shrinkage and selection operator LR model. Of these, those with P > 0.05 were excluded, and 19 predictors with P < 0.05 were selected for the univariate LR analysis (Table 2). After adjusting the model based on expert opinion, a total of 17 predictors were decided.
| Feature | Univariate logistic regression | Multivariate LR | ||
| OR (95%CI) | P value | OR (95%CI) | P value | |
| Age, yr | 2.64 (2.06–3.37) | < 0.001 | 2.68 (1.99-3.61) | < 0.001 |
| BNP | 1.53 (1.34–1.75) | < 0.001 | ||
| CRP | 2.01 (1.68–2.40) | < 0.001 | ||
| D-dimer | 1.13 (1.06–1.20) | < 0.001 | ||
| Albumin | 0.37 (0.29–0.48) | < 0.001 | 0.62 (0.46–0.83) | 0.001 |
| Hcv_ab | 1.005 (0.999–1.011) | 0.083 | ||
| AST | 1.50 (1.29–1.75) | < 0.001 | ||
| CysC | 1.32 (1.19–1.46) | < 0.001 | ||
| APTT | 1.80 (1.45–2.23) | < 0.001 | ||
| Cr | 1.10 (1.05–1.16) | < 0.001 | 1.11 (1.06–1.15) | < 0.001 |
| Myoglobin | 1.64 (1.44–1.88) | < 0.001 | ||
| CK | 1.12 (1.05–1.20) | < 0.001 | ||
| PCT | 1.008 (1.003–1.014) | < 0.001 | ||
| Tp_ab | 1.001 (0.999–1.003) | 0.231 | ||
| BUN | 1.70 (1.44–1.99) | < 0.001 | ||
| PT | 1.65 (1.36–1.99) | < 0.001 | ||
| LAC | 0.81 (0.68–0.98) | 0.012 | ||
| LDH | 2.36 (1.91–2.93) | < 0.001 | 2.09 (1.61–2.7) | < 0.001 |
| Hgb | 0.69 (0.58–0.82) | < 0.001 | ||
| GLU | 1.28 (1.15–1.44) | < 0.001 | 1.16 (1.02–1.31) | 0.023 |
| HBeAg | 1.010 (0.920–1.108) | 0.156 | ||
| HBsAg | 0.999 (0.995–1.003) | 0.577 | ||
| NLR | 1.90 (1.61–2.25) | < 0.001 | 1.27 (1.06–1.51) | 0.008 |
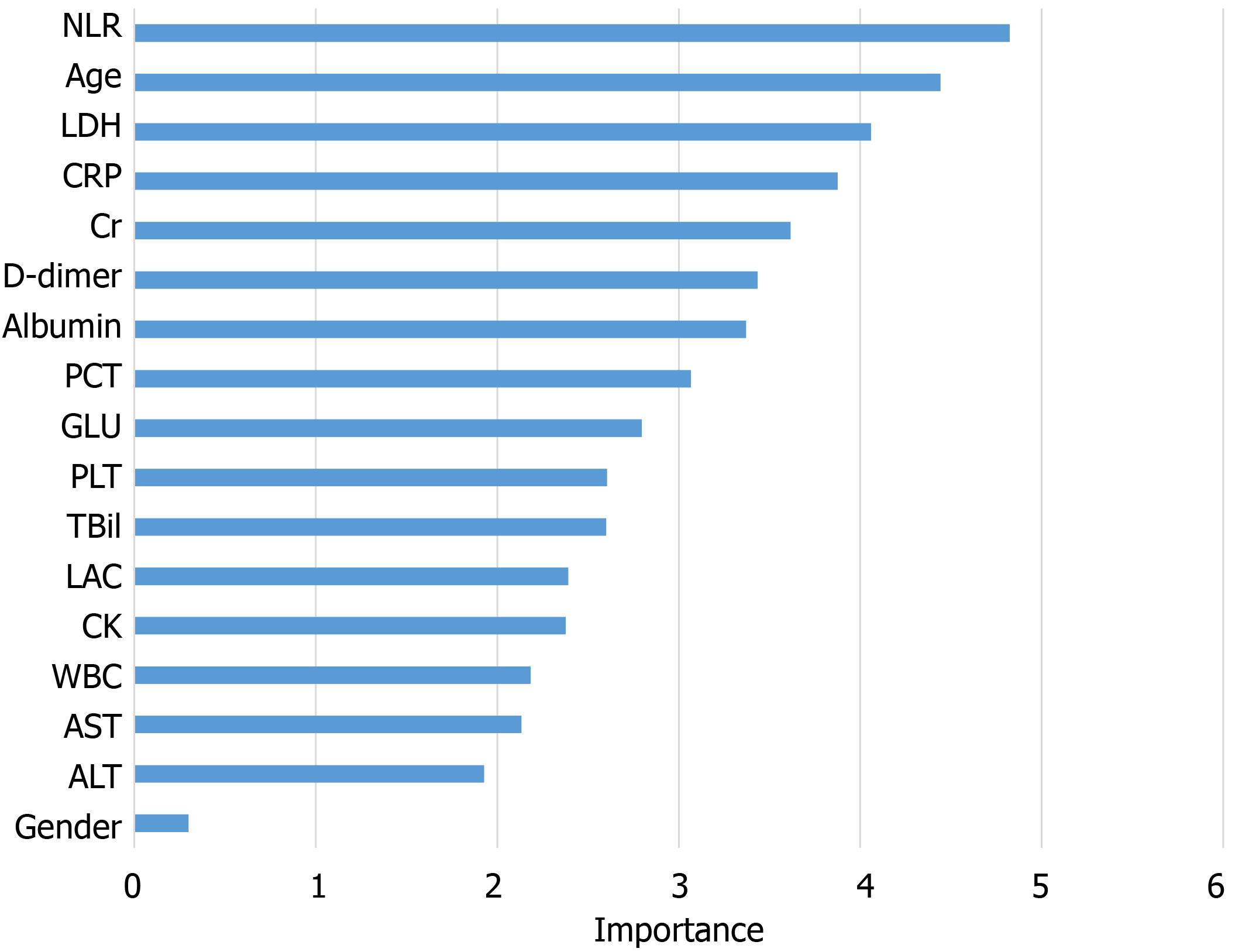
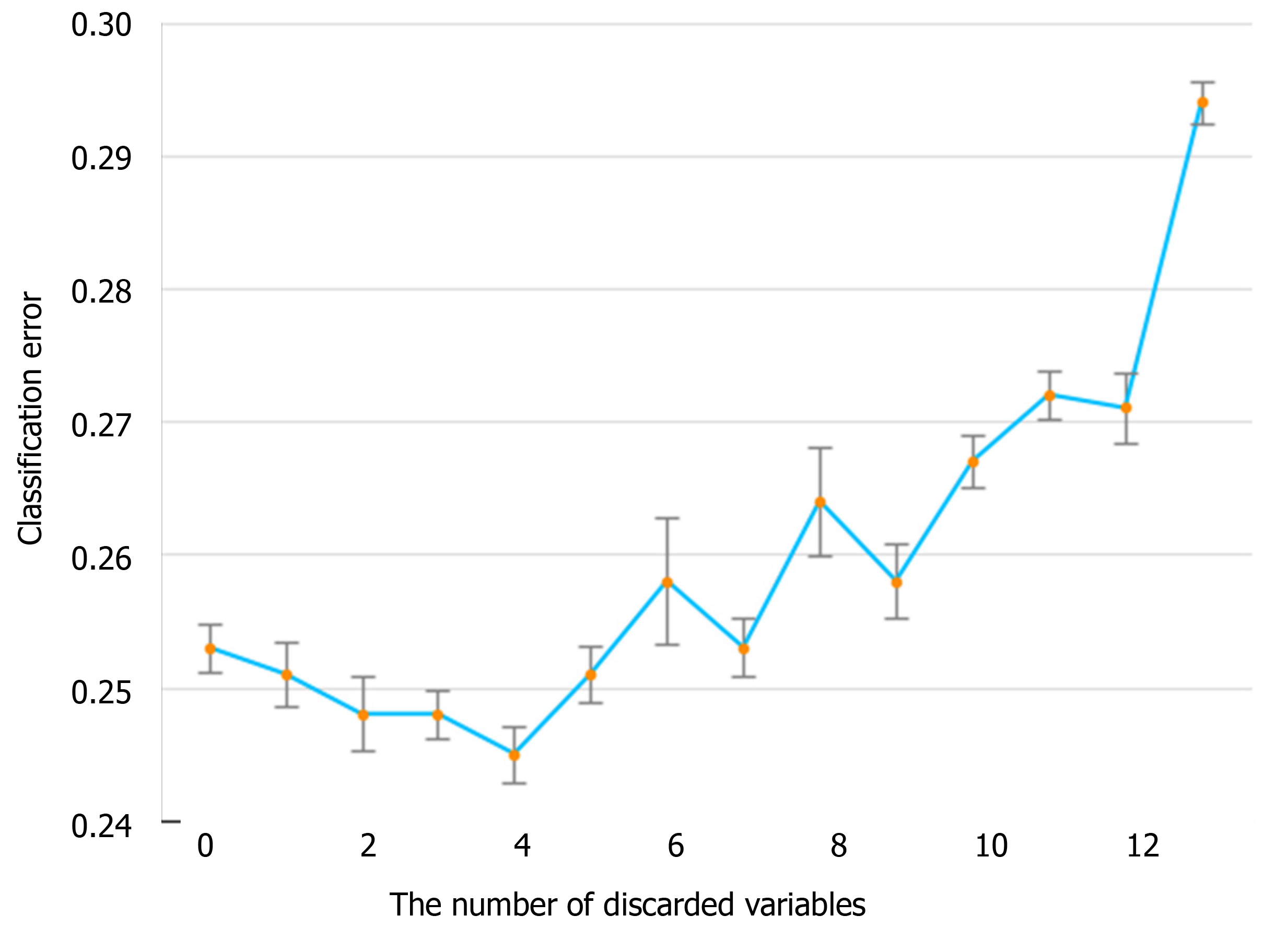
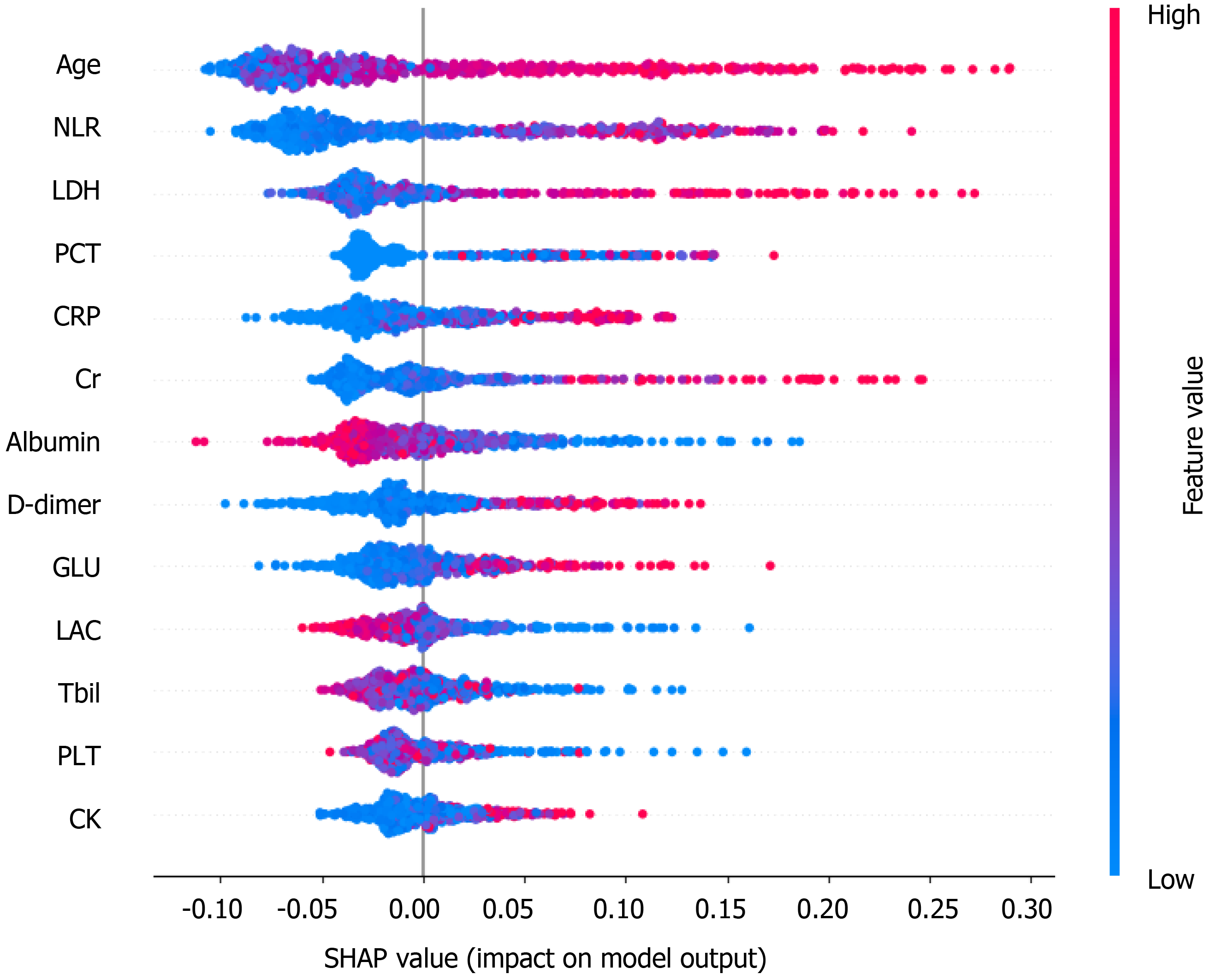
The information gain of each variable and its importance were calculated and ranked based on entropy and out-of-bag error. Figure 3 depicts the relative importance of each of the features. Variables were dropped from the bottom of the list, starting from the variable that was deemed least important and progressing in ascending order. The least classification error was obtained when gender, alanine transaminase, aspartate aminotransferase and white blood cell were dropped from the prediction model. Hence, these four features were removed. Figure 4 shows the relationship between the number of discarded variables and classification error. After all, a final of 13 predictors were selected. SHapley Additive exPlanations value was calculated to explain the output of the RF model (Figure 5). It connected optimal credit allocation with local explanations using the classic Shapley values from game theory and their related extensions[35]. In the next step, six predictors [neutrophil-to-lymphocyte ratio (NLR), age, lactate dehydrogenase, creatinine, glucose and albumin] with P < 0.05 were selected to build the multivariate LR model (Table 2). The personalized nomogram was then used to show the probability of ICU admission (Figure 6).
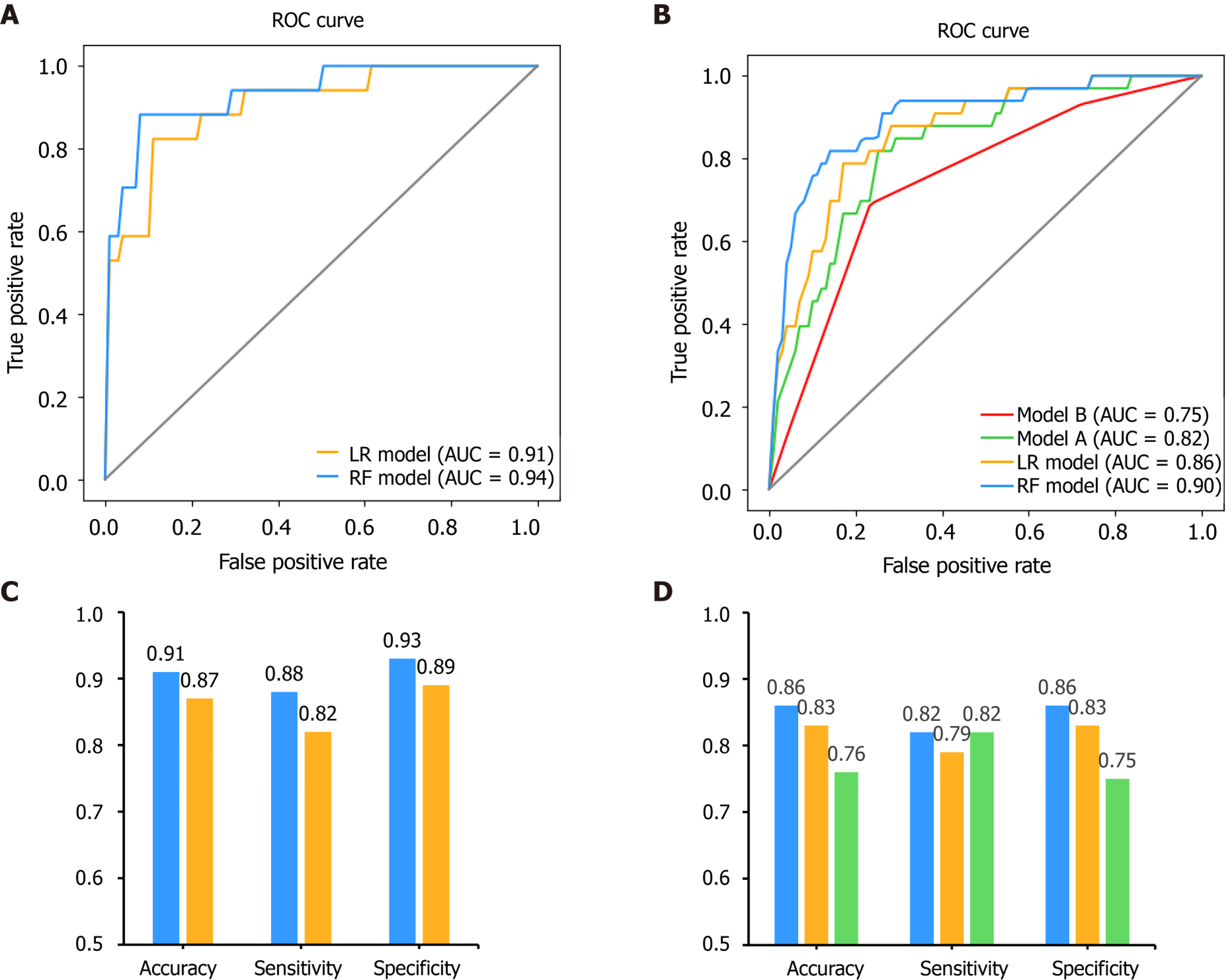
After feature selection, two predictive models, LR and RF, were built on the basis of the selected variables. For internal validation, the area under the receiver operating characteristic curve for the RF model was found to be 0.94, which was higher than that for the LR model at 0.91 (P = 0.111, DeLong’s test). The accuracy, sensitivity and specificity for the RF model were 91%, 88% and 93%, respectively, higher than those for the LR model (87%, 82%, and 89%, respectively) (Figure 7).
Moreover, we compared our results with those of previously published methods in the external validation dataset as well (Figure 7). The area under the receiver operating characteristic curves for the RF, LR, model A and model B models were 0.90, 0.86, 0.82 and 0.75, respectively. The DeLong’s test between these models was: P = 0.135 (RF vs LR), P = 0.004 (RF vs model A), P = 0.006 (RF vs model B), P = 0.333 (LR vs model A), P = 0.045 (LR vs model B) and P = 0.185 (model A vs model B). The accuracy for the RF, LR and model A models were 86%, 83% and 76%, respectively. The sensitivity was 82%, 79% and 82%, respectively. The specificity was 86%, 83% and 75%, respectively. Since model B showed the lowest performance in this validation set, only the predictive performance of LR, RF and model A were compared for the following experiments.
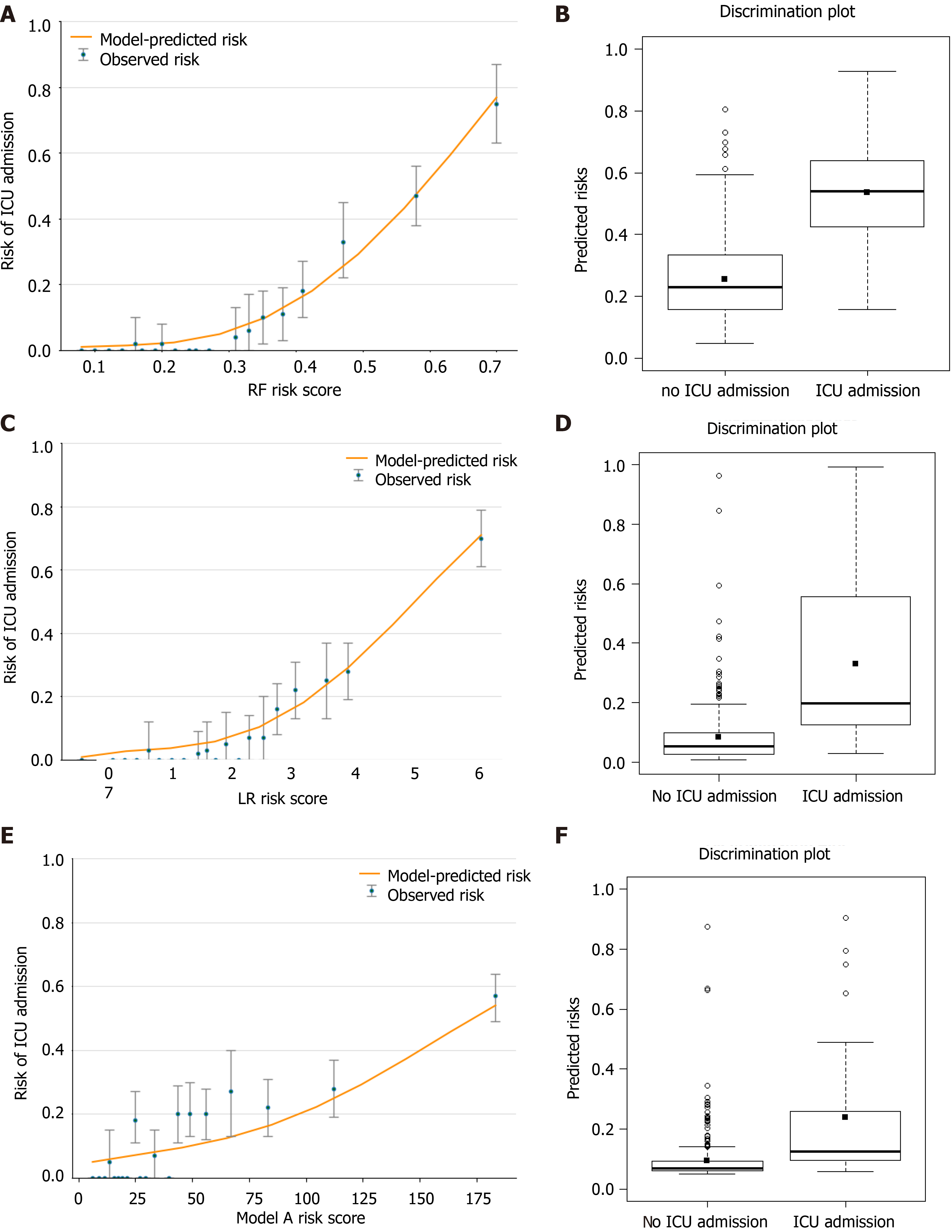
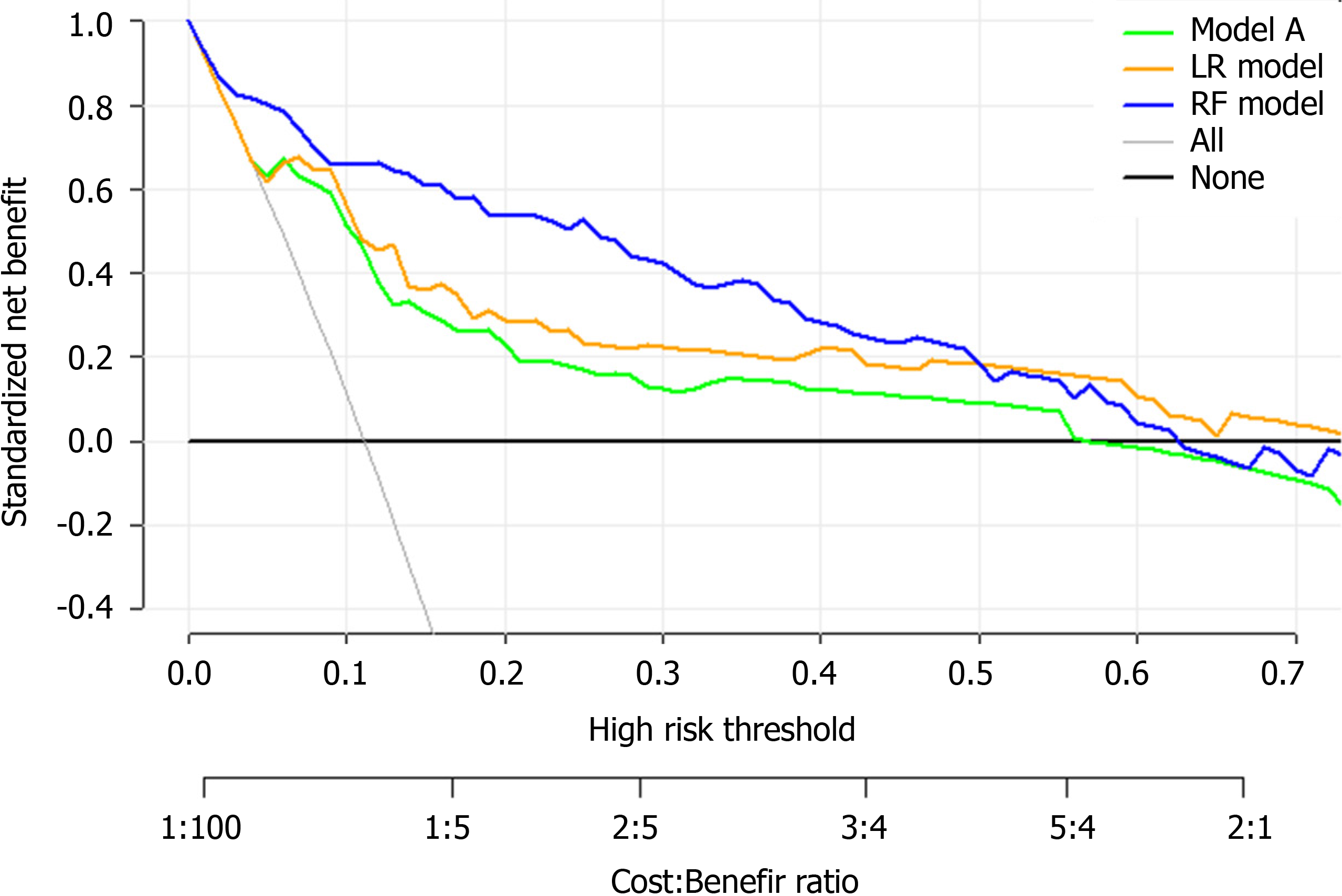
Figure 8A, 8C and 8E shows the observed risk of ICU admission vs model-predicted risk in groups based on the calculated model risk score. The overestimation and underestimation in the probability range of ICU admission risk were evident from the plots. Model A underestimated risk in patients with a predicted risk less than 30%. The RF and LR models show a perfect calibration. The RF risk score demonstrated an excellent ability to categorize patients in separate risk strata. Figure 8B, 8D and 8F shows the differences in the predicted probability values between ICU admission and no ICU admission using each of the prediction models. The discrimination slope for the RF, LR and model A models were 0.281, 0.246 and 0.143, respectively. The plots indicated a lack of fit for the model A.
We further calculated the risk of each individual in the entire testing cohort and divided all patients into three groups based on the risk cut-off at 95% sensitivity and 95% specificity[36]. Then we computed the NRI with RF as the second model, and LR and model A as the first models. In this scenario, a positive NRI value would indicate that the RF model has better discriminative ability compared to the other models, while a negative value would indicate the converse. Table 3 summarizes the results of this analysis.
| Initial model | Updated model | Predicted probability according to initial model | Statistics | |||||
| < 13% | 13%-20% | > 20% | % Reclassified | NRI (95%CI) | P value | |||
| Model A | RF | < 13% | 189 | 46 | 9 | 23 | 0.363 (0.148-0.579) | < 0.001 |
| 13%-20% | 6 | 12 | 5 | 48 | ||||
| > 20% | 2 | 13 | 14 | 52 | ||||
| LR | < 13% | 208 | 17 | 19 | 15 | 0.246 (0.029-0.463) | 0.026 | |
| 13%-20% | 14 | 3 | 6 | 87 | ||||
| > 20% | 8 | 4 | 17 | 41 | ||||
| LR | RF | < 13% | 194 | 36 | 0 | 16 | 0.167 (-0.024-0.357) | 0.087 |
| 13%-20% | 3 | 17 | 4 | 29 | ||||
| > 20% | 0 | 18 | 24 | 43 | ||||
Finally, DCA was used to facilitate the comparison between different prediction models. As shown in Figure 9, the DCA graphically shows the clinical usefulness of each model based on a continuum of potential thresholds for major high risk and the standardized net benefit of using the model to stratify patients relative to assuming that there were no ICU admission patients. As shown from our results, the standardized net benefit yielded by the models developed in this study was larger across the major high-risk range compared with model A.
Due to the rapidly expanding number of patients and the limited resources in the ICU, prediction models for patients with COVID-19 are crucial in clinical decision-making and medical resource micro-allocation. In the present study, a training cohort of 681 COVID-19 patients were recruited from Wuhan Third Hospital. A risk prediction model was successfully established to assess the chance of ICU admission based on the lab results obtained at the time of hospital admission. Furthermore, we performed an external validation on a total of 296 confirmed COVID-19 patients from Shenzhen Third People’s Hospital. Comparing with the recent published methods on the same validation data, our results revealed that the newly developed model (RF) exhibited relatively better discriminatory power, and the external verification was also satisfactory. In addition, our model showed a better discriminatory power in diverse populations from hospitals of different levels with varying death rates and varying baseline physical conditions, indicating that our models that were developed in the current study can be applied to a wide variety of settings.
Meanwhile, when creating a new prediction model, we recommend selecting predictors based on previous literature and expert opinion, rather than in a purely data-driven way[8]. In this case, we developed a mixed-knowledge feature selection process, including machine-selection and clinicians’ knowledge, together with previous published predictors[21]. Although the more information used during the developing step, the better performance the models would be, we would like to limit the number of the predictors while achieving similar performance in order to ease the user experience. Several studies have shown that lung imaging can help assess disease severity in COVID-19[37], which is one of the clinical diagnostic criteria. However, our predictive model was able to achieve good results by using only the biochemical indicators obtained on the first day of admission, thereby reducing the physical strain and economic burden on patients and governments.
Furthermore, based on the selected variables, two clinical predictive models were built. The LR model used only six of the selected variables but performed better than the other two published methods[33,34] on our external validated dataset. One step further, in order to improve the predictive ability of the mode, a more sophisticated machine learning method, RF, was introduced in our model building step.
The predictive performance of the models built in the present study were carefully evaluated and calibrated. As we all know, poorly calibrated models will underestimate or overestimate the outcome of interest, while an excellent model will show strong calibration for different groups of patients. A model with adequate calibration by predicted risk strata will provide useful information for clinical decision making[28]. In our results, visual representation of the relationship between predicted and observed values were shown to evaluation calibration. Discrimination and calibration results show that the RF model demonstrated an excellent ability to categorize patients in separate risk strata and the values predicted by the model agree with the observed values, which indicated that both the RF and LR models performed better than other published methods, and the RF model performed the best. Furthermore, when comparing the performance of all the three models, the reclassification result revealed that the RF model resulted in the reclassification of a large number of patients, and a positive NRI value indicated that the RF model performed better than the other models.
The following six variables were the most important in prediction of a risk of ICU admission among COVID-19 patients, in decreasing order of importance: NLR, age, lactate dehydrogenase, C-reactive protein, creatinine, D-dimer and albumin.
The NLR represents inflammation and is a known indicator of the systemic inflammatory response[38]. Yang et al[38] stated that elevated NLR could be considered an independent biomarker of indicating poor clinical outcomes in the outcome following COVID-19. Age was the second most important factor in the model, and age has been very well known as an important biomarker of poor clinical outcomes in the context of COVID-19. Lactate dehydrogenase and C-reactive protein have also been found to be associated with poorer outcomes such as respiratory failure in COVID-19 patients[39]. These results are in agreement with the findings of our study.
Finally, the clinical model developed in this study may be able to assist medical professionals to identify high-risk patients at their first assessment in settings where medical resources are limited. Based on the results of the DCA, the standardized net benefit was the highest with the RF model across the major high-risk range. This model can aid doctors infer the likely course of COVID-19 at an early stage so that they can guide the patients toward more appropriate treatments. Therefore, patients that are more likely to develop a severe case of COVID-19 can get close attention and high-level treatments in advance.
The present study has some limitations. First, the participants included patients who tested positive for COVID-19 in Wuhan; therefore, studies across larger areas need to be carried out to further verify our findings. Second, any medications taken prior to hospital admission and the time interval between hospital admission and disease onset could have affected the data records. Third, we did not analyze some data points, e.g., the body mass index and viral load, which are potential risk factors of infection severity, in our study. Despite these limitations, our predictive models yielded good discriminatory power when we verified the models in a heterogeneous population. Fourth, some data that may be critical to a patient’s prognosis, such as mechanical ventilation data, were not collected in this study. However, in China, treatment for COVID-19 among all hospitals is carried out in line with the National Health Commission of China guidelines[26]. Here, we devised predictive models using patient information obtained from tests done at admission. In the future, research should include repeated measures data to identify whether any temporal changes in clinical indicators are better able to predict disease prognosis in COVID-19.
In the present study, we used the first day of laboratory results to build a model for the early prediction of the need for ICU admission among patients diagnosed with COVID-19 infection. Upon external verification, the discriminatory powers exhibited by our predictive models were relatively satisfactory. The models developed in this study can aid high-risk patients to achieve early intervention and provide guidance to ensure the rational allotment of medical resources.
The novel coronavirus disease 2019 (COVID-19) pandemic is a global threat caused by the severe acute respiratory syndrome coronavirus-2.
The development of a prognostic model is crucial to address the problem of micro-allocation of scarce healthcare resources in the face of a pandemic.
To develop and validate a risk stratification tool for the early prediction of intensive care unit admission among COVID-19 patients at hospital admission.
We selected 13 of 65 baseline laboratory results and developed a risk prediction model with the random forest algorithm. A nomogram for the logistic regression model was built based on six selected variables.
The accuracy, sensitivity and specificity for the random forest model were 91%, 88% and 93%, respectively, higher than those for the logistic regression model. The area under the receiver operating characteristic curve of our model was much better than those of two other published methods (0.90 vs 0.82 and 0.75).
Our model can identify intensive care unit admission risk in COVID-19 patients at admission, who can then receive prompt care, thus improving medical resource allocation.
In the future, research should include repeated measures data to identify whether temporal changes in clinical indicators are better able to predict disease prognosis in COVID-19.
Manuscript source: Unsolicited manuscript
Specialty type: Medical informatics
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A, A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Balakrishnan DS, Islam SMRU, Naswhan AJ, Patel J, Ssekandi AM S-Editor: Wu YXJ L-Editor: Filipodia P-Editor: Xing YX
| 1. | Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J Med Virol. 2020;92:401-402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1670] [Cited by in F6Publishing: 1693] [Article Influence: 423.3] [Reference Citation Analysis (0)] |
| 2. | Hui DS, I Azhar E, Madani TA, Ntoumi F, Kock R, Dar O, Ippolito G, Mchugh TD, Memish ZA, Drosten C, Zumla A, Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264-266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2274] [Cited by in F6Publishing: 1861] [Article Influence: 465.3] [Reference Citation Analysis (0)] |
| 3. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32663] [Cited by in F6Publishing: 28482] [Article Influence: 7120.5] [Reference Citation Analysis (3)] |
| 4. | Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13871] [Cited by in F6Publishing: 12369] [Article Influence: 3092.3] [Reference Citation Analysis (1)] |
| 5. | Topçu HO, Kokanalı K, Güzel AI, Tokmak A, Erkılınç S, Ümit C, Doğanay M. Risk factors for adverse clinical outcomes in patients with tubo-ovarian abscess. J Obstet Gynaecol. 2015;35:699-702. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Sun Y, Koh V, Marimuthu K, Ng OT, Young B, Vasoo S, Chan M, Lee VJM, De PP, Barkham T, Lin RTP, Cook AR, Leo YS; National Centre for Infectious Diseases COVID-19 Outbreak Research Team. Epidemiological and Clinical Predictors of COVID-19. Clin Infect Dis. 2020;71:786-792. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 150] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 7. | Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514-523. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6159] [Cited by in F6Publishing: 5253] [Article Influence: 1313.3] [Reference Citation Analysis (0)] |
| 8. | Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, Bonten MMJ, Dahly DL, Damen JAA, Debray TPA, de Jong VMT, De Vos M, Dhiman P, Haller MC, Harhay MO, Henckaerts L, Heus P, Kammer M, Kreuzberger N, Lohmann A, Luijken K, Ma J, Martin GP, McLernon DJ, Andaur Navarro CL, Reitsma JB, Sergeant JC, Shi C, Skoetz N, Smits LJM, Snell KIE, Sperrin M, Spijker R, Steyerberg EW, Takada T, Tzoulaki I, van Kuijk SMJ, van Bussel B, van der Horst ICC, van Royen FS, Verbakel JY, Wallisch C, Wilkinson J, Wolff R, Hooft L, Moons KGM, van Smeden M. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1689] [Cited by in F6Publishing: 1607] [Article Influence: 401.8] [Reference Citation Analysis (0)] |
| 9. | Colombi D, Bodini FC, Petrini M, Maffi G, Morelli N, Milanese G, Silva M, Sverzellati N, Michieletti E. Well-aerated Lung on Admitting Chest CT to Predict Adverse Outcome in COVID-19 Pneumonia. Radiology. 2020;296:E86-E96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 316] [Cited by in F6Publishing: 304] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 10. | Das AK, Mishra S, Gopalan SS. Predicting community mortality risk due to CoVID-19 using machine learning and development of a prediction tool. 2020. [Cited in This Article: ] |
| 11. | Singh K, Valley TS, Tang S, Li BY, Kamran F, Sjoding MW, Wiens J, Otles E, Donnelly JP, Wei MY, McBride JP, Cao J, Penoza C, Ayanian JZ, Nallamothu BK. Evaluating a Widely Implemented Proprietary Deterioration Index Model Among Hospitalized COVID-19 Patients. medRxiv. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Zhang H, Shi T, Wu X, Zhang X, Guthrie B. Risk Prediction for Poor Outcome and Death in Hospital In-Patients with COVID-19: Derivation in Wuhan, China and External Validation in London, UK. SSRN Electronic J. 2020;. [Cited in This Article: ] |
| 13. | Bai X, Fang C, Zhou Y, Bai S, Liu Z, Xia L, Chen Q, Xu Y, Xia T, Gong S. Predicting COVID-19 Malignant Progression with AI Techniques. Social ence Electronic Publishing. [Cited in This Article: ] |
| 14. | Gong J, Ou J, Qiu X, Jie Y, Chen Y, Yuan L, Cao J, Tan M, Xu W, Zheng F, Shi Y, Hu B. A Tool for Early Prediction of Severe Coronavirus Disease 2019 (COVID-19): A Multicenter Study Using the Risk Nomogram in Wuhan and Guangdong, China. Clin Infect Dis. 2020;71:833-840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 318] [Article Influence: 79.5] [Reference Citation Analysis (0)] |
| 15. | Huang J, Cheng A, Lin S, Zhu Y, Chen G. Individualized prediction nomograms for disease progression in mild COVID-19. J Med Virol. 2020;92:2074-2080. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 16. | Rubin D, Chan-Tack K, Farley J, Sherwat A. FDA Approval of Remdesivir - A Step in the Right Direction. N Engl J Med. 2020;383:2598-2600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 125] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 17. | Gostin LO, Salmon DA, Larson HJ. Mandating COVID-19 Vaccines. JAMA. 2021;325:532-533. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 79] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 18. | Kent DM, Paulus JK, Sharp RR, Hajizadeh N. When predictions are used to allocate scarce health care resources: three considerations for models in the era of Covid-19. Diagn Progn Res. 2020;4:11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Wang F, Nie J, Wang H, Zhao Q, Xiong Y, Deng L, Song S, Ma Z, Mo P, Zhang Y. Characteristics of Peripheral Lymphocyte Subset Alteration in COVID-19 Pneumonia. J Infect Dis. 2020;221:1762-1769. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 670] [Cited by in F6Publishing: 753] [Article Influence: 188.3] [Reference Citation Analysis (0)] |
| 20. | Breiman L. Bagging Predictors. Machine Learning. 1996;24:123-140. [DOI] [Cited in This Article: ] |
| 21. | Bellou V, Tzoulaki I, Evangelou E, Belbasis L. Risk factors for adverse clinical outcomes in patients with COVID-19: A systematic review and meta-analysis. 2020 Preprint. Available from: medRxiv:2020.2005.2013.20100495. [DOI] [Cited in This Article: ] |
| 22. | Lieske JC, Chawla L, Kashani K, Kellum JA, Koyner JL, Mehta RL. Biomarkers for acute kidney injury: where are we today? Clin Chem. 2014;60:294-300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Parikh RB, Schwartz JS, Navathe AS. Beyond Genes and Molecules - A Precision Delivery Initiative for Precision Medicine. N Engl J Med. 2017;376:1609-1612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 24. | Chang CT, Tsai TY, Liao HY, Chang CM, Jheng JS, Huang WH, Chou CY, Chen CJ. Double Filtration Plasma Apheresis Shortens Hospital Admission Duration of Patients With Severe Hypertriglyceridemia-Associated Acute Pancreatitis. Pancreas. 2016;45:606-612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Collins GS, Reitsma JB, Altman DG, Moons KGM; members of the TRIPOD group. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD Statement. Eur Urol. 2015;67:1142-1151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 275] [Cited by in F6Publishing: 258] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 26. | Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: The Hosmer-Lemeshow test revisited. Crit Care Med. 2007;35:2052-2056. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 476] [Cited by in F6Publishing: 584] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 27. | Bertolini G, D'Amico R, Nardi D, Tinazzi A, Apolone G. One model, several results: the paradox of the Hosmer-Lemeshow goodness-of-fit test for the logistic regression model. J Epidemiol Biostat. 2000;5:251-253. [Cited in This Article: ] |
| 28. | Alba AC, Agoritsas T, Walsh M, Hanna S, Iorio A, Devereaux PJ, McGinn T, Guyatt G. Discrimination and Calibration of Clinical Prediction Models: Users' Guides to the Medical Literature. JAMA. 2017;318:1377-1384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 632] [Cited by in F6Publishing: 789] [Article Influence: 112.7] [Reference Citation Analysis (1)] |
| 29. | Pencina MJ, D'Agostino RB Sr, D'Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157-72; discussion 207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4325] [Cited by in F6Publishing: 4789] [Article Influence: 299.3] [Reference Citation Analysis (0)] |
| 30. | Pirracchio R, Petersen ML, Carone M, Rigon MR, Chevret S, van der Laan MJ. Mortality prediction in intensive care units with the Super ICU Learner Algorithm (SICULA): a population-based study. Lancet Respir Med. 2015;3:42-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in F6Publishing: 203] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 31. | Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 596] [Cited by in F6Publishing: 849] [Article Influence: 53.1] [Reference Citation Analysis (0)] |
| 32. | Kerr KF, Brown MD, Zhu K, Janes H. Assessing the Clinical Impact of Risk Prediction Models With Decision Curves: Guidance for Correct Interpretation and Appropriate Use. J Clin Oncol. 2016;34:2534-2540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 338] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 33. | Wu S, Du Z, Shen S, Zhang B, Yang H, Li X, Cui W, Cheng F, Huang J. Identification and Validation of a Novel Clinical Signature to Predict the Prognosis in Confirmed Coronavirus Disease 2019 Patients. Clin Infect Dis. 2020;71:3154-3162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 34. | Shi Y, Yu X, Zhao H, Wang H, Zhao R, Sheng J. Host susceptibility to severe COVID-19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit Care. 2020;24:108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 346] [Cited by in F6Publishing: 334] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 35. | Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, Nair B, Katz R, Himmelfarb J, Bansal N, Lee SI. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat Mach Intell. 2020;2:56-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2808] [Cited by in F6Publishing: 1357] [Article Influence: 339.3] [Reference Citation Analysis (0)] |
| 36. | Liang W, Yao J, Chen A, Lv Q, Zanin M, Liu J, Wong S, Li Y, Lu J, Liang H, Chen G, Guo H, Guo J, Zhou R, Ou L, Zhou N, Chen H, Yang F, Han X, Huan W, Tang W, Guan W, Chen Z, Zhao Y, Sang L, Xu Y, Wang W, Li S, Lu L, Zhang N, Zhong N, Huang J, He J. Early triage of critically ill COVID-19 patients using deep learning. Nat Commun. 2020;11:3543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 146] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 37. | Li J, Yu X, Hu S, Lin Z, Xiong N, Gao Y. COVID-19 targets the right lung. Crit Care. 2020;24:339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 38. | Yang AP, Liu JP, Tao WQ, Li HM. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharmacol. 2020;84:106504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 605] [Cited by in F6Publishing: 551] [Article Influence: 137.8] [Reference Citation Analysis (0)] |
| 39. | Poggiali E, Zaino D, Immovilli P, Rovero L, Losi G, Dacrema A, Nuccetelli M, Vadacca GB, Guidetti D, Vercelli A, Magnacavallo A, Bernardini S, Terracciano C. Lactate dehydrogenase and C-reactive protein as predictors of respiratory failure in CoVID-19 patients. Clin Chim Acta. 2020;509:135-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 128] [Article Influence: 32.0] [Reference Citation Analysis (0)] |