Abstract
The coronavirus disease (COVID-19) outbreak reported in China in December 2019 has spread throughout the world. The WHO declared it as a pandemic in March 2020. The pandemic severely affected public health and the global economy. Many studies conducted on the coronavirus have helped us to elucidate its pathogenicity and pathophysiology. However, it is important to study the behavior of the pathogen in the environment to develop effective control measures. While studying the persistence and transmission of viruses in drinking water and wastewater systems, a low concentration of coronavirus and its nucleic acids have been detected in municipal wastewaters. This could be due to their high susceptibilities to degradation in aqueous environments. Epidemiological study on coronaviruses in wastewater will serve two purposes, i.e., in early detection of outbreak and in identifying asymptomatic carriers. In such cases, the epidemiological study will help in early detection of the presence of the virus in the community. Secondly, it will help in knowing if there are asymptomatic carriers, as such people do not show any signs of symptoms but shed the viruses in feces. The present review focuses on the epidemiological surveillance of wastewater for coronaviruses, as in recent years these are increasingly causing global pandemics. In this review we have discussed, the four pertinent areas of coronavirus study: (1) occurrence of coronavirus in wastewater, (2) wastewater based epidemiological surveillance of coronaviruses, (3) epidemiological surveillance tools used for detection of coronaviruses in sewage, and (4) persistence and sustainability of coronaviruses in wastewater.
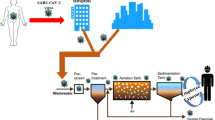
Graphical abstract

Similar content being viewed by others
Introduction
The pneumonia outbreak caused by severe acute respiratory syndrome coronavirus-2 (SARS-COV-2) was first reported in Wuhan, China, in December 2019. By March 2020, as the infectious disease spread across several countries, the World Health Organization (WHO) declared it as a pandemic (World Health Organization, 2020). The total number of recorded infected cases and morbidity is escalating, with the pandemic severely affecting public health and the global economy (Ali et al., 2020). The severity of the COVID-19 pandemic is unprecedented in the modern era. People with pre-existing morbidities are highly susceptible to COVID-19 infection. Physicians and healthcare workers are at high risk as they are constantly exposed to the coronavirus during patient care and treatment. Similarly, healthcare workers performing diagnostic procedures and those administering therapeutics are also at high risk (CDC, 2007). Therefore, the pandemic caused by the novel coronavirus-2 is a challenge to the scientific and medical community.
Coronaviruses are classified as belonging to family Coronaviridae, subfamily Orthocoronavirinae, and order Nidovirales. Based on phylogenetic clustering, the Coronaviridae family is classified into four genera and includes Alpha, Beta, Gamma, and Delta coronaviruses (Colford et al., 2006). Coronaviruses were identified for the first time in the 1930s during the investigation of acute respiratory infections in domesticated chickens. The causal organism was identified as the infectious bronchitis virus (IBV). Later in the 1960s, human coronaviruses were identified in patients suffering from common cold and named as human coronavirus-229E and human coronavirus-OC43 (Leung et al., 2003). In recent years, the coronavirus SARS-COV in 2003, MERS-COV in 2012, and SARS-COV-2 in 2019 have been identified as causative agents of serious respiratory tract infections (Figs. 1). COVID-19 causing novel coronavirus-2 can infect different organs, apart from causing severe acute respiratory problems and pneumonia (La Rosa et al., 2020).
The viruses are referred to as “coronavirus” because electron microscopy image shows the presence of large bulbous projections resembling a crown or a solar corona on the virus surface. The projections or spikes observed on the surface of the virus are proteinaceous and termed as peplomers. Coronavirus are large enveloped pleomorphic spherical particles of 120 nm size. The envelope is a lipid bilayer made up of membrane (M), envelope (E), and spike (S) structural proteins. Nucleocapsid consists of multiple copies of protein attached to positive-sense RNA, and the conformation is seen as continuous beads on a string. The genome of coronaviruses is approximately 27–34 kb. In comparison to other RNA viruses, coronavirus is considered to be the largest virus (Daughton, 2020).
Coronaviruses HCoV-229E, SARS-CoV, HCoV-OC43, MERS-CoV, HCoV-N, and HCoV-HKU1 infect humans. Whereas coronaviruses porcine enteric diarrhea CoV (PEDV), bat coronaviruses (BatCoV), feline infectious peritonitis virus (FIPV), bovine coronavirus (BCoV), and transmissible gastroenteritis virus (TGEV) infect wildlife, pets, and livestock (La Rosa et al., 2020). Coronaviruses capable of causing both respiratory and gastrointestinal infections are listed in Table 1.
The virus infection is initiated when it comes in contact and binds to the receptors present on the complementary host cells. The proteases secreted by host cell cleave and activate the receptors attached to the viral spike. Viruses enter the host either by endocytosis or through direct fusion with the cell membranes. The genome of the virus, i.e., the RNA, is released into the cytoplasm. It uses the host cell ribosomal machinery for its translation and synthesis of nonstructural proteins required for its assembly. Further, it is transmitted from the respiratory droplets that are shed by infected patients during sneezing and coughing (Daughton, 2020).
Concerns over reemergence of the SARS coronavirus have led to increased research on vaccine development. Certain coronavirus-related diseases such as infectious bronchitis in birds and canine and feline coronavirus have been the target of vaccine production (Cavanagh, 2003). Currently, vaccines based on 12 types of platforms are at pre-clinical and clinical level trials against SARS-CoV-2 (Fig. 2). Various vaccine types being evaluated include nucleic acid-, viral vector-, virus-, and protein subunits, of which over 70 vaccines use protein subunit platform, 30 use non-replicating viral vector platforms, and 29 use RNA platforms. Currently, there are 66 vaccines in clinical trials, with 17 trials in phase I, 23 trials in phases I–II, 6 trials in phase II, and 20 trials in phase III. The trials of four candidates were halted. In phase III trials, some of the COVID 19 vaccines showed up to 95% efficacy in preventing symptomatic COVID-19 infections (Mellet & Pepper, 2021). Currently, the various vaccine candidates undergoing phase III clinical trials and approved for emergency use are listed in Table 2. Among the pharmacological therapeutic strategies, Remdesivir (GS-5734) a broad-spectrum antiviral drug was the first antiviral agents to be evaluated against SARS-CoV-2. Clinical trials are currently testing its safety and efficacy for COVID-19 treatment in humans. Fostamatinib, an inhibitor of the spleen tyrosine kinase, plays a role in adaptive immune receptor signaling, innate immune recognition, and platelet function. This FDA approved drug is currently in phase II clinical trials for COVID-19. Similarly, clinical trials are conducted to evaluate the efficacy and timing of corticosteroid treatment for COVID-19. Convalescent plasma obtained from recovered COVID-19 patients containing naturally produced antibodies have also been explored for clinical administration, but these warrant standardization of protocols. Among the antibodies, the first synthetic one for clinical use has been REGN-COV2 antibody cocktail developed by Regenron (Izda et al., 2020). With the emergence of SARS-CoV-2, both therapeutic approaches and a vaccine solution are being investigated relentlessly to control COVID-19 morbidity and mortality.
Occurrence of coronavirus in wastewater
An individual excretes a large number of microorganisms in feces and urine which are carried away in sewage. Enteric pathogens capable of surviving in water cause waterborne diseases and are transmitted through fecal–oral-route (Guan et al., 2003). Several viruses, bacteria, and protozoans are identified as enteric pathogens as they are transmitted by consumption of drinking water contaminated with wastewater. These pathogens can also spread through contaminated surfaces or from person to person contact (Dayaram et al., 2017). Infected individuals shed millions of virus particles in their feces or body fluids, which eventually enter sewage systems. Effective management of human excreta in municipal waste is the primary step for the prevention of the pathogen from spreading into the environment (Garcia-Aljaro et al., 2019).
The presence of enteric viruses in aquatic environments is due to sewage discharge, which is considered to be the primary source of water contamination. Commonly occurring pathogenic enteric viruses in wastewater include norovirus, enterovirus, adenovirus, astrovirus, and rotavirus. The gastroenteritis outbreaks across the world have been mainly attributed to direct or indirect exposure of humans to treated sewage water containing persistent pathogens or their spores. The enteric viruses can replicate only in human hosts and cause a wide range of infections in humans with diverse symptoms (Parasidis et al., 2013).
In 1975, coronaviruses were first observed using electron microscopy in the feces of a person with the gastroenteric infection. Investigation of severe acute respiratory syndrome (SARS) outbreak in 2003 revealed that the transmission was limited to people in close contact with patients. However, the possible transmission from the wastewater system could not be ruled out as patients excreted coronavirus and RNA of these viruses were found in the stools samples in Amoy Gardens, Hong Kong. Similarly, coronavirus responsible for Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak in 2013 caused atypical presentations including gastroenteritis in infected patients (Peiris et al., 2003). In the current situation, the major focus of the global scientific community is on understanding the pathogenic behavior and environmental persistence of the novel coronavirus-2.
In the pandemic affected countries, scientists are analyzing sewage water for the presence of novel coronavirus. Such surveillance of wastewater helps in calculating the total number of infected cases prevalent in a community (Le Poder, 2011). A single waste treatment plant contains wastewater from more than one million people (Mallapaty, 2020). Therefore, successful detection of virus or their gene fragments will be useful in calculating the approximate number of people infected in a geographic area. Thus, the time and resources required for testing every individual can be reduced. Surveillance of wastewater will help in identifying infections at initial stages and set up an early warning system for outbreak detection (Mallapaty, 2020). Such alert and response systems will ensure effective control of the outbreak and prevent it from escalating into a pandemic like situations.
In recent years, there is a considerable increase in the studies on the occurrence of coronaviruses in water and wastewater environments in many countries (Table 3). In the future, wastewater-based epidemiology will be a potential tool for disease monitoring and for devising control strategies during epidemic events. Quantifying and calculating the pathogenic viruses in wastewater will help in determining the degree of treatment required to reduce the health risk due to infections (Yuan et al., 2020).
Wastewater-based epidemiological surveillance of coronaviruses
Wastewater-based epidemiology (WBE) is a new concept. It gives detailed and comprehensive information about health risks in communities. It is based on extraction, detection, inspecting, and elucidating chemical or biological compounds (also known as biomarkers) from wastewater matrix (Sims & Kasprzyk-Hordern, 2020). The concept of WBE was introduced by Daughton in 2001 for analyzing the drug residue in sewage water (Daughton, 2001). Further, the concept was adopted for extracting and quantifying cocaine in both the surface water and wastewater (Zuccato et al., 2005). Similarly, for effective management of current situations such as COVID-19 pandemic, WBE techniques can provide unbiased information of microbial risk in wastewaters (Kasprzyk-Horden et al., 2014).
Monitoring of diseases and rapid reliable information of disease outbreak in a community can be identified by utilizing WBE as a complementary surveillance technique (Sims & Kasprzyk-Hordern, 2020).The use of WBE has given good results in drug consumption monitoring and population-wide exposure. However, its application in the current pandemic like situation needs to be discussed (Choi et al., 2019; Daughton, 2018). The identification of potential biomarkers which could be pathogenic bacteria and viruses or their constituents could be helpful in monitoring wastewater for detecting the presence and potential spread of infectious diseases in the community (Sims & Kasprzyk-Hordern, 2020).
The genetic material (RNA) of pathogenic organisms serves as a potential biomarker, and their detection in feces (< 1 × 101–106.5) has been used as an indicator of severe acute respiratory syndrome (SARS-CoV) in the community (Poon et al., 2004). The detection of such biological markers (pathogenic DNA/RNA) in influent sewage water implies that the disease is circulating within the community. The viral pathogens present in the aquatic environment show a significant threat for humans due to their high rate of mutation and adaptation ability inside the human cells. Thus, in such cases, WBE surveillance techniques can be used for the detection of these viruses (Woolhouse & Gowtage-Sequeria, 2005). Carducci and his team worked on understanding virological risk assessment in the entry and exit point of the wastewater plants. Their study reveals that treated wastewater contains infectious human viral pathogen. The finest indicator for inactivation of viruses in recycled water seems to be adenovirus followed by somatic coliphages (Carducci et al., 2009).
The concept of the WBE approach is promising with potential results in the hepatitis A and norovirus outbreak (Hellmér et al., 2014). The WHO guidelines about monitoring polio in the wastewater matrix are good proof for the success of WBE in analyzing poliovirus circulation within the population (World Health Organization, 2003). Similar research about persistence and surveillance of the coronavirus in aquatic environments will give a complete picture regarding its potential transmission risk in the community (La Rosa et al., 2012).
Many studies that have analyzed municipal wastewater from affected communities have shown the presence of the pathogenic virus. Wastewater-based epidemiology can help in detecting the prevalence of pathogenic viruses in the wastewater treatment plant as the effluent contains viruses excreted from both the asymptomatic and symptomatic individuals in the community (Sinclair et al., 2009; Xagoraraki & O'Brien, 2020). Wastewater-based epidemiology tool has been successfully used to caution and notify the early disease outbreaks of enteric viruses including norovirus, hepatitis A virus, and poliovirus resulting in the effective public health interventions (Hellmér et al., 2014; Asghar et al., 2014). The estimation and concentration of viral RNA in wastewaters have shown a high range of correlation with the epidemic status of the population linked to the wastewater network (Ahmed et al., 2020a, b).
WBE techniques help in giving desired information regarding environmental exposure and health status of a community. Reasonable and replicable methods are used for the detection of viral pathogens in contaminated water (Sims et al., 2020). Pandemic situations demand adequate detection and a survey of virus distribution in the environment. WBE can be used for early prediction and to point out the viral hotspots to protect the public from risk (Venugopal et al., 2020). Detection of viral pathogens in sewage water could help public health officials to initiate early intervention measures including quarantine and other preventive steps. Such timely identification of the virus arrival in a community can result in prompt action towards the control of outbreak and reduce the health and economic damage caused by novel viruses including SARS-COV-2 and other re-emerging viruses (World Health Organization, 2020a).
The emerging and remerging disease outbreaks and its surveillance have become an integral component of public health risk assessment. Therefore, disease monitoring is one of the best ways to analyze the disease outbreak. Water fingerprinting is also considered to be one of the best methods in WBS. Here, the extraction, detection, and analysis of biomarkers reflect the potential health risk in water and wastewater (Sims & Kasprzyk-Hordern, 2020). In Australia, WBE has been used as a potential tool for SARS-CoV surveillance and monitoring. The presence of COVID-19 in wastewater was confirmed using molecular methods including RT-qPCR and sequencing techniques (Ahmed et al., 2020a, b). Similar studies on the possible transmission of coronaviruses from sewage have been conducted using advanced molecular techniques such as PCR. Wang and his team in China found nucleic acid of SARS-CoV in sewage before disinfection in Beijing Hospital and 309th Hospitals of the Chinese People’s Liberation Army. They adopted an electropositive filter media to concentrate virus from sewage, followed by semi-nested RT-PCR and gene sequencing to confirm its presence. On analyzing disinfected sewage samples from the 309th Hospital, the presence of a virus was confirmed. The study revealed that the virus can survive for 14 days in sewage at 40 °C and for 2 days at 20 °C. The SARS-CoV could survive for 8 days, though the inactivation process was adopted (Wang et al., 2005a, b, c). Southwestern Greece wastewater plant was examined for 2 years to evaluate and detect the viral circulation in the city. PCR techniques were adopted to survey the viral strains, such as hepatitis A viruses, noroviruses, adenoviruses, human polyomaviruses, and hepatitis E viruses in sewage water. The study revealed the distribution and frequency of pathogenic viral strains in the community. Around 87.5% of the virus was identified in the wastewater sample. The result confirmed the presence of both DNA and RNA viruses in wastewater treatment plants (Kokkinos et al., 2011). Thus, WBE when equipped with molecular tools such as PCR can be used for rapid and early detection of a potential viral outbreak in the community. Dutch scientists detected the presence of COVID-19 in sewage waters collected from Amsterdam Schiphol airport in Tilburg, Netherlands. This was the first study reporting the detection of SARS-COV-2 in sewage in the current outbreak situation. Thus, surveillance of wastewater can be a sensitive tool for monitoring community spread of the virus, especially when the coronavirus has low prevalence (Mallapaty, 2020).
Similarly, surveillance for variants in wastewater could be used to rapidly detect emergence of variants of CoV-2 and monitor their spread in a community. A number of new SARS-CoV-2 variants with high number of mutations have been reported across the world. These variants have been classified by the Centers for Disease Control and Prevention (CDC) as variants of interest, variants of concern, and variants of high consequence based on certain attributes. Three new variants B.1.1.7, 501Y.V2 (B.1.351), and P.1 (B.1.1.28.1) are of foremost importance and are classified as variants of concern (VoC) due to their high transmissibility and antibody escape. These VoC lineages have gained attention as they are rapidly spreading at high efficiency worldwide. Large-scale genomic screening study has led to the identification of Lineage B.1.1.7 (23 mutations with 17 amino acid changes) in the UK. Through similar surveillance studies, lineage B.1.351 (23 mutations with 17 amino acid changes) was first documented in South Africa followed by emergence of lineage P.1 (approximately 35 mutations with 17 amino acid changes) in Brazil (Abdool Karim & de Oliveira, 2021). These variants have N501Y mutation which changes the amino acid in the receptor-binding domain of the spike protein. The key concerns of the new variants are their viral transmissibility, disease severity, reinfection rates, and vaccine effectiveness. Lineages B.1.351 and P.1 have shown increased resistant to antibody neutralization (Wang et al., 2021). Recently, few more SARS-CoV-2 variants, B.1.427 and B.1.429 first detected in California, and B.1.617, B.1.617.1, B.1.617.2, and B.1.617.3 detected in India have shown more transmissibility than other variants. Therefore, the spread of these variants in the community should be monitored to know their transmission dynamics and take appropriate measures to keep them in check. Variants surveillance done through whole genome sequencing of clinical samples is expensive and time-consuming. Therefore, genome sequencing of viral RNA isolated from sewage can rapidly detect and identify any sequence variation (Izquierdo-Lara et al., 2021). Thus, deep sequencing initiatives in sewage samples can help detect new and rare mutations from mixtures of SARS-CoV-2 lineages and identify VoC in sewage and subsequently their presence in the community (Heijnen et al., 2021).
Qualitative microbial risk assessment of wastewater
The adaptation of water safety plans is a very important guideline for maintaining the quality of water. Qualitative microbial risk assessment (QMRA) is an answerable model for water treatment monitoring. The model assesses the health-based target and requires wastewater monitoring for safe drinking water (Smeets et al., 2010).
More research and studies on valid and reproducible techniques for the perception of viral pathogens in the wastewater is required. This will help in regulating the scope of contamination of the aquatic environment and understanding the correlation between viral contaminations with aquatic environmental factors. QMRA technique helps in understanding the use of water quality data, pathogenic-specific characteristics, prevalence, and exposure data. This will help in providing awareness of health risks associated with wastewater system and upgrade outbreak control methods (La Rosa et al., 2012). QMRA is a helpful tool in detecting health risks associated with identifying pathogens and understanding its relation in the environment. It includes field data and laboratory experiments to measure and quantify the disease in a specific exposed environment. Previously, QMRA has been successfully used in detecting food-borne disease outbreak and now implied for application in waterborne disease caused by enteric pathogens (Bentham & Whiley, 2018). The guidelines regarding QMRA by the World Health Organization include important steps such as (1) hazard identification, (2) dose response, (3) exposure assessment, and (4) risk characterization (Ashbolt, 2004). QMRA is an ideal means of predicting and analyzing the microbial risks in a community (Bentham & Whiley, 2018). The Federal-Provincial-Territorial Committee on Drinking Water (CDW) of Canada documented the risks associated with the circulation of enteric viruses in potable water. The minimum 4-log inactivation level was required to remove pathogenic viruses in drinking water sources. Characterization of risk related to water sources is required for implementing necessary disinfection methods for controlling the circulation of the pathogenic viruses in water sources (CDC, 2017). QMRA for pathogenic enteric viruses present in drinking water followed by the committee on drinking water in Canada has been discussed in Table 4. In the present COVID-19 scenario, QMRA in WWTPs can be developed by taking into considerations factors such as shedding of CoV-2 into raw wastewater, concentration of CoV-2 in raw and treated wastewater, aerosolized water inhaled by workers, effect of humidity and temperature on viability of aerosolized coronaviruses, and frequency and duration of exposure (Dada & Gyawali, 2021). In three COVID-19 scenarios, QMRA was used to estimate the risk of infection in WWTP workers based on viral loads and infectivity of CoV-2 in sewage affluent. Such QMRA study is useful for risk management decisions and to plan emergency response (Zaneti et al., 2021).
Detection methods for coronavirus in wastewaters
Wastewater-based epidemiology consists of four main steps which include sampling of wastewater, storage of sample, concentration step, extraction, and detection (Alygizakis et al., 2021). Sampling is one of the most crucial steps as it can affect the entire analysis. As seen for the analysis of SARS-CoV-2 in wastewaters, though composite sampling is recommended WBE technique, yet grab sampling has been used more often. However, parameters including flow rate, volume of wastewater, and temperature are not considered in this sampling technique. This limitation of grab sampling can be avoided by using 24-h flow/time composite samples. Sample volume should also be given importance as low volume can lead to low recovery and result in false-negative results. Thus, increased volume can produce signals even if recovery is low (Hart & Halden, 2020). During WBE, samples have to be stored at temperature of – 20 °C to inactivate the bacterial activity and for degradation of contaminants of emerging concern. But this temperature is not favorable for CoV-2 genetic material. Further, freezing and thawing can lead to RNAses degrading CoV-2 RNA in wastewater, and the loss in viral load could impact further analysis analysis (Alygizakis et al., 2021). The storage protocol for samples and genetic materials has to be optimized at the earliest. A number of concentration methods are used for enriching CoV-2 from wastewater. But these methods lack use of ideal external control standards having properties similar to SARS-CoV-2. On comparative analysis of seven concentration methods for recovery of CoV-2 in wastewater, absorption-extraction method proved to be the most rapid and effective. Adding MgCl2 to pre-filtered wastewater improved recovery when using electronegative membrane. Amicon Ultra-15 filter used in ultrafiltration showed acceptable functioning performance. But sample volume and the filter type are also important for analysis. Use of PEG gave low yields and increased concentration of PCR inhibitors. Though pre-centrifugation removed larger particles and debris, it lowered recovery as pellet absorbs genetic material (Ahmed et al., 2020a, b). Isolation of CoV-2 RNA from concentrated extracts is performed by commercially available kits as they include lysis and inhibitor which guarantees high-quality, ready-to-use RNA in highly complex matrices. However, the yield of SARS-CoV-2 virus recovered, and RNA isolated must be quantified. This could be done by adding non-target RNA sequence after concentration or addition of the lysis solution (Medema et al., 2020). For the detection of CoV-2, a number of assays for virus detection differing in sensitivity and specificity using various pairs of specific primers and probes are reported. However, Centers for Disease Control and Prevention recommended protocol, primers and probes are highly used. In the WBE studies, the first PCR assays for CoV-2 was developed against the E-gene and the N-gene. Later PCR assays targeting RdRP and ORF1ab were also reported (La Rosa et al., 2020). The limitation of PCR assays are possibilities of false-negative and false-positive results due to any of the reasons such as errors in sampling, low viral load in samples, and degradation of the RNA. CoV-2 detection in wastewaters has been confirmed using the “gold standard” RT-qPCR which uses one-step RT-qPCR with efficient cDNA synthesis and real-time PCR in a single tube (Rimoldi et al., 2020). Apart from RT-qPCR, ultrasensitive assays using nested PCR and droplet digital PCR (ddPCR) have been used in WBE studies. Nested PCR uses two sets of primers and thus increases sensitivity by reducing the non-specific binding. ddPCR improves the limit of detection and accuracy in the detection of CoV-2 even in samples with low viral load. It gives absolute quantification of virus genome copy numbers without the need for external calibration (Liu et al., 2020). However, it is expensive as it requires the use of high cost instruments and reagents. Genome sequencing methods have been used to eliminate false-positive PCR results as wastewater is a complex matrix with contaminants affecting analysis. Though sequencing is not favorable for screening of massive samples in clinical settings, yet the next-generation sequencing has potential in WBE studies (Westhaus et al., 2020).
Nucleic acid-based RT-qPCR and metagenomics for detection of CoV-2 in wastewater
A number of wastewater-based epidemiological surveillance on COVID-19 has been conducted in different parts of the world. In this section, the techniques adopted by the researchers worldwide for the sample collection, processing, and detecting the viral genetic material have been reviewed. The merits and limitations of the methods used have been discussed. The findings and implications of these studies have been summarized in Table 5. The primers and probes used in RT-qPCR for detection of CoV-2 in wastewater samples have been listed in Table 6.
SARS-CoV-2 RNA was detected in domestic wastewater of cities and a main airport during the early stages of the COVID-19 epidemic in the Netherlands. RT-qPCR was performed with the primers for N1–N3 set and E gene. During the initial days of infection, viral RNA was detected in sewage at a concentration of 2.6 –30 gene copies per mL. In one particular sampling location, N3 was detected in sewage 6 days prior to the reporting of first case. It was observed that with the increase in COVID-19, the viral RNA detected for N1–N3 genes by qRT-PCR assay increased to 790–2200 gene copies per mL. The detection of the CoV-2 RNA in sewage before the onset of COVID-19 and during the initial days of infection clearly indicates the usefulness of sewage surveillance as a sensitive tool to monitor the prevalence of virus in the community (Heijnen et al., 2021). In a WBE study in nine municipal wastewater treatment plants in Germany, samples were processed and analyzed for a set of CoV-2-specific genes and pan-genotypic gene sequences using RT-qPCR. The study showed the presence of CoV-2 genetic traces in different raw wastewaters. Sanger sequencing confirmed the specificity of the assay and origin of the coronavirus (Westhaus et al., 2020). In an Australian WBE study, SARS-CoV-2 RNA was concentrated from wastewater using two types of automated sampling techniques — refrigerated autosampler or a submersible in-situ high frequency autosampler and grab sampling techniques. Viruses were concentrated by direct RNA extraction from electronegative membranes. The viral RNA copies were enumerated using RT-qPCR, and viral RNA copy numbers were used to estimate infected individuals via Monte Carlo simulation. A median range of 171 to 1,090 infected persons in the catchment was estimated in agreement with clinical observations. N_Sarbeco RT-qPCR followed by Sanger and Illumina sequencing showed that the N_Sarbeco assay is more sensitive than the NIID_2019-nCOV_N assay as the LOD of the assay was 8.3 copies/reaction (Corman et al., 2020).
A study conducted for a period of 74 days in a WWTP located in Bozeman, Montana (USA), showed that WBE with genome sequencing can be used to monitor viral prevalence in the community and simultaneously help in genotyping circulating viral strains in a community. Untreated wastewater samples were collected using autosampler. Prior to RNA extraction, they were filtered and concentrated using ultrafiltration with spin concentrators. qRT-PCR reaction was performed with extracted RNA using primer pairs for nucleocapsid (N) gene from SARS-CoV-2. qRT-PCR study provided a real-time measure of viral prevalence in the community as samples tested positive for SARS-CoV-2 mimicked with COVID-19 cases in the community. Phylogenetic analyses using nanopore genome sequencing of 14,970 SARS-CoV-2 genomes from 74 different countries showed that the Bozeman wastewater SARS-CoV-2 sequence differed from the Wuhan-Hu-1/2019 reference sequence but was closely related to viral strains from California and Victoria, Australia. Thus, such a surveillance study helps in understanding COVID-19 epidemiology and tracing virus spread patterns. The limitations of the study were the nanopore sequencing which has an error rate of ∼ 10–15% resulting in detection of genotypes with > 10–15% representation. In this study, a single method was used for the concentration of virus from sample, and this rules out comparative analysis of alternative efficient concentration methods (Nemudryi et al., 2020).
In a WBE study conducted in Water Resource Recovery Facility (WRRF) in Michigan, SARS-electropositive NanoCeram column filters were used to isolate CoV-2 from wastewater samples. The capacity of the filter was 40–70 L of influent with a flow rate of 11.3 L/min. CoV-2 was isolated from these filters and quantified by qRT-PCR. These filters could be stored at 4 °C until elution and until 48 h after sampling. The elution techniques is simple, wherein cartridge used for filtration is submerged in beef extract solution for 1 min. Further, beef extract eluate is magnetically stirred and then centrifuged at 3,800 rpm for 15 min. The pellet contains virus and is dissolved in Na2HPO4 to prevent virus loss. A two-step RT-qPCR was performed with primers and probe targeting the N1 gene of SARS-CoV-2. It detected and quantified SARS-CoV-2 in all the samples tested at a concentration of 104–105 genomic copies/L. Thus, a modified procedure in sample processing and virus recovery proved to be efficacious (Miyani et al., 2020).
In WBE in urban treatment facility in Massachusetts, two methods were used to detect CoV-2 RNA. Initially, Sanger sequencing was used to validate the S gene. It was followed by RT-qPCR with primer and probe sets targeting the N1, N2, and N3 loci to detect N gene (57 to 303 copies per ml of sewage). Raw sewage precipitated with polyethylene glycol 8000 (PEG) and 0.2-μm-filtered sewage were comparatively analyzed for viral enrichment. The former was found to give strongest and consistent results for RNA extraction. Thus, a simple viral enrichment protocol can be used for viral identification. The study also reported that pasteurization of the sample for 90 min at 60 °C as a first step ensured the safety of the protocol. The protocol followed here is simple and less expensive avoiding usage of expensive chemicals and commercial kits for viral RNA extraction. The viral titers obtained through this study were higher than clinically confirmed cases. This could be due to less loss in viral titer during sample processing or RNA extraction (Wu et al., 2020). Similarly, a simple procedure was used in WBE study in a Wastewater Treatment Plant (WWTP) at Ahmedabad, India. ORF1ab, N and S genes of SARS-CoV-2 were found in the influent with the estimated maximum concentration of 3.5 × 102 copies/L. The high SARS-CoV-2 genetic loading in the wastewater concurred with corresponding increase in the active COVID-19 cases in the city. However, no genes were detected in the effluent collected during the same period. The study asserts the need for monitoring temporal changes in SARS-CoV-2 RNA concentrations in wastewater (Kumar et al., 2020). Such a study was conducted for comparison of SARS-CoV-2 viral load in wastewater influents, treated effluents, and untreated wastewater in the United Arab Emirates (UAE). The samples were collected for over 24 h and thermally deactivated, followed with ultrafiltration, RNA extraction, and viral quantification using RT-qPCR. Viral load in wastewater influents ranged from 7.50E + 02 to over 3.40E + 04 viral gene copies/L, and in untreated wastewater samples, it was between 2.86E + 02 and over 2.90E + 04 gene copies/L. However, none of the treated effluents tested positive for the virus which implies efficient degradation of SARS-CoV-2 and safety of the treated water for re-use. Thus, the study not only emphasizes wastewater testing in monitoring an outbreak at the community level but also helps in assessing the safety of treated effluent (Hasan et al., 2021).
The use of an aluminum hydroxide adsorption-precipitation concentration method for CoV-2 in wastewater sample has been validated. Influent, secondary, and tertiary effluent water samples in six WWTPs in Spain were monitored for CoV-2 using the said concentration method. Quantification of CoV-2 RNA in untreated wastewater by RT-qPCR targeting three regions of the nucleocapsid N gene, led to estimation of 5.4 ± 0.2 log10 genomic copies/L. None of the tertiary water samples tested positive. The surveillance data revealed CoV-2 RNA in sewage even before the first case was reported by local or national authorities. Thus, WBE could be used an early indicator of the infection in a community. The limitation in the studies were the discrepancies observed in the RT-qPCR for N1, N2, and N3 assays for several samples which could be due to variation in analytical sensitivity, detection of possible false positive samples for N3, and inhibitory effect of matrix. The authors have suggested exclusion of N3 primers/probe set to limit false-positive results and use of digital RT-qPCR for more sensitive estimation of SARS-CoV-2 in wastewaters (Randazzo et al., 2020).
Concentration of samples by ultracentrifugation was found to be more efficient than filtration in a study conducted in WWTP in Paris. Ultracentrifugation was easier to perform and showed better CoV-2 recovery rate than filtration as approx. 2.5 106 UG/L was detected. The study adopted a specific RT-qPCR where initially E gene was detected and quantified. Further, positive results were confirmed by amplification of viral RdRp gene. The time-course monitoring of viral load in WW was compared with total number of COVID-19 cases. The observed delay between epidemiological curves in humans and viral RNA detected in WW was attributed to various reasons including timing and temporal kinetics of viral RNA shedding in feces. The study also indirectly proved relevant reduction of virus transmission during lockdown. Thus, real-time integrated WBE of CoV-2 helped in identifying stages of epidemics, virus circulation in community, and effectiveness of lockdowns and such barrier measures (Wurtzer et al., 2020).
Sewage surveillance for SARS-CoV-2 in WWTP in Italy was analyzed with influent wastewater samples collected before and after the onset of the epidemic. Both nested RT-PCR and real-rime RT-PCR assays was used for detecting and quantifying viral genes. The limit of detection (LOD) of virus in wastewater was found to be to 5.6 × 104 genome copies/L. The study showed that SARS-CoV-2 was already circulating in various geographic locations of Italy before the first Italian COVID-19 case was documented. The report highlights the importance of WBE surveillance for early warning of virus circulating in the community. Similarly the study also documents the use of modified protocol for the surveillance of poliovirus in sewage and concentration by two-phase (PEG-dextran) separation method (La Rosa et al., 2021). This study is also an improvisation of an earlier study on the first detection of SARS-CoV-2 in wastewater in Italy (La Rosa et al., 2020). In the previous study, no positive results were obtained by real-time RT-qPCR. Therefore, in this study, a newly designed real-time RT-(q)PCR assay was evaluated, and nested RT-PCR targeting the ORF1ab region was used. The study stresses that sewage being complex matrix may always not give complicit results for assays developed mainly for clinical samples (La Rosa et al., 2021).
Similar novel nested RT-PCR approach was used to detect the presence of CoV-2 RNA in wastewater in South East England. The RT-qPCR assay targeting five different regions of the viral genome resulted in improved sensitivity. Further, NGS analysis of PCR products showed single nucleotide polymorphisms at five selected nucleotide positions. This helped to detect co-circulating virus variants, specifically prevalent in England. Further, changes in sequences with time consistent with the increase in presence of Spike protein G614 pandemic variant were also identified. Thus, RT-qPCR clubbed with NGS helps in detection of the presence of CoV-2 variants in wastewater which further implies its prevalence in the community (Martin et al., 2020).
RT-qPCR and RT-ddPCR assays were used to detect and quantify CoV-2 in primary treated sewage samples in a WWTP in Canada. The genes targeted were N1 and N2 gene regions of the viral RNA. Signal inhibition in certain samples was observed while using RT-ddPCR, whereas RT-qPCR shows higher frequency of detection of N1 and N2 gene regions. The study also showed that choice of the sample also matters as higher viral load was detected in clarified sludge than grit solids. The study also made use of pepper mild mottle virus (PMMoV) as a biomarker for normalization of CoV-2 signal. PMMoV, a fecal indicator, can be used for monitoring when there is low incidence of SARS-CoV-2 infection in communities (D'Aoust et al., 2021).
Two techniques were used for concentration of SARS-CoV-2 in wastewater samples in Sweden. Milk powder and NanoCeram filter were used for flocculation and adsorption for sampling the process. NanoCeram filter was more sensitive for CoV-2 detection. Milk powder may not adsorb enveloped viruses, whereas the non-enveloped SARS-CoV-2 nucleocapsids were adsorbed. But this is not a problem in case of NanoCeram filter as both encapsidated and non-encapsidated forms adsorbed to it. Based on variation in the viral peaks with regular temporal intervals, it was implied that CoV-2 may have a cluster spread. A correlation was observed between increase in concentrations of CoV-2 RNA detected in wastewater and newly hospitalized COVID-19 infected patients as CoV-2 detected in wastewater preceded virus detection in patient clinical samples. Such studies are useful to rapidly identify local spread of the virus (Saguti et al., 2021).
Viral metagenomics is an advanced study that has found potential application in epidemiological surveillance. The use of metagenomics tool for detecting SARS-CoV-2 in wastewater in a study in Spain has resulted in the detection of variants circulating in the community. The study used 40 grab samples collected from 14 WWTPs in Spain. Influent samples were concentrated by aluminum-based adsorption precipitation method. SARS-CoV-2 nucleic acid was detected by RT-qPCR targeting three genomic regions, i.e., the N1 region, E gene, and IP4 gene. ARTIC protocol version 3 was used for genomic sequencing of CoV-2 from wastewater samples. Sequencing libraries were built and sequenced on the Illumina MiSeq platform by paired-end reads. For the variant analysis, SARS-CoV-2 isolate Wuhan-Hu-1 (MN908947.3) was used as reference genome. Through the study, 238 nucleotide substitutions, 6 deletions, and nucleotide variants were detected in the spike glycoprotein. Three novel variants in the spike gene and six new variants in the spike glycoprotein were reported for the first time. The metagenomic approach helps in analyzing large number of genomes and aids in detecting low-frequency variants circulating in a community. Metagenomic sequencing of sewage can be used for monitoring new mutations and variants of SARS-CoV-2 (Perez-Cataluna et al., 2021). A similar study conducted using NGS analysis of sewage samples has stressed the surveillance of wastewater to monitor for viral diversity in a geographic location and emergence of novel mutations. The 4 primer–probe sets targeting the N1–N3 and E gene were used for RT-qPCR quantification, while CoV-2-specific multiplex PCR was used for nanopore sequencing. Next-generation sequencing and phylogenetic analysis of sewage samples collected from the Netherlands and Belgium revealed the presence of prevalent clades 19A, 20A, and 20B and several novel mutations in the CoV-2 genome. Nanopore and Illumina NGS analysis was useful in studying the diversity of CoV-2 in sewage as 51 novel mutations in sewage consensus sequences were detected that were not previously reported. Low-frequency variants analysis showed 8 novel mutations not previously observed in either the Netherlands–Belgium or global datasets. Thus, NGS analysis of wastewater could be used to study CoV-2 diversity in a community. Genome sequencing of sewage could be either used independently or with clinical surveillance to observe for changes in viral diversity and clinically relevant mutations (Izquierdo-Lara et al., 2021).
Droplet digital RT-PCR to detect SARS-CoV-2
In a study conducted in Wuhan, detection of CoV-2 in urban wastewater treatment plants and medical wastewater was compared using two techniques, qPCR and droplet digital PCR (ddPCR) assay, and by targeting ORF1ab, N gene and E gene. CoV-2 RNA was detected in all the tested samples by either qPCR or ddPCR. The LoDs of ddPCR were 2 copies/reaction for all three targets. The detection rates of qPCR were 50% for ORF1ab in ddPCR; it was 100%. qPCR for one of the concentrated sample showed no presence of N gene, while ddPCR assay detected CoV-2 RNA in the same sample. Thus, ddPCR seems to have high sensitivity for the complex wastewater samples (Zhou et al., 2021).
RT-ddPCR has also been used in wastewater samples for specific detection of SARS-CoV-2 variants such as mutation N501Y. N501Y and wild-type could be simultaneously detected from a mixture of CoV-2 RNA in raw sewage samples in a study conducted in the Netherlands. Detection of N501Y wastewater aligned with B.1.1.7 as causative CoV-2 in COVID-19 cases. The variants with B.1.1.7, B.1.351, and P.1 lineages are known to contain N501Y mutation. The mutation is known to influence spike protein binding to its host receptor. In this study, digital droplet RT-PCR primers were used to amplify 80 bp fragment of the spike gene where mutation leads to the N501Y amino acid change in the spike protein. FAM-labeled probe that binds to N501Y mutation and HEX-labeled probe to bind wild-type CoV-2 sequence were used. In ddPCR technique, PCR occurs in separate self-contained droplets which help to identify mutant fragments at low frequencies amidst wild-type fragments. Thus, using ddPCR rare mutations can be detected and closely related sequences distinguished. Application of RT-ddPCR for wastewater assessment can help in monitoring presence of any mutants in the community (Heijnen et al., 2021).
Thus, different concentration and extraction methods have been used to determine the viral gene in the wastewater matrix (Hjelmso et al., 2017; Sherchan et al., 2020). Molecular techniques such as digital PCR (dPCR) and next-generation sequencing provide efficient and rapid information about the presence of different microbial pathogens in samples. The metagenomics technique could have potential applications in epidemiological studies of WWTP during pandemic situations (Sims & Kasprzyk-Hordern, 2020). Advanced WBE techniques can overcome the limitations associated with conventional techniques. Another potential technology that could be used to detect the pathogen in a sample is the CRISPR (clustered regularly interspaced short palindromic repeats). It is a powerful, highly specific and versatile tool and provides unprecedented control over genome editing. CRISPR/Cas9 has been used for detection of certain human viruses and has potential applications in novel therapeutic approaches (White et al., 2015). Paper-based tests have been developed for rapid and easy detection of COVID-19 from wastewater for predicting the infection level in the community. Here, the virus present in sewage water is used as a biomarker. The preloaded reagents in the device detect the presence of nucleic acid from SARS-CoV-2, which can be visualized and does not require any readout device. Such techniques are useful for identifying asymptomatic carriers present in the community for prevention and early warning (Mao et al., 2020). Serological and molecular techniques are also useful in quantifying viruses in the sewage water matrix. Indirect immunofluorescence and flow cytometer method has been used to detect infectious human retroviruses (Reslova et al., 2017). Some of the other methods used for virus detection and analysis in the aquatic environment are electron microscopy, nucleic acid sequence-based amplification, fluorescent microscopy, microarray techniques, and flow cytometry. A combination of different methods also gives good results for detection, such as PCR with plaque-forming tests and protein microarray technology with atomic force microscopy (Hryniszyn & Skonieczna, 2013). The advantages and limitations associated with the techniques used for the detection of viruses in wastewater are listed in Table 7.
Persistence and sustainability of coronaviruses in wastewater
Coronaviruses are not a major problem for wastewater and water industries as they are present in low concentrations. They are highly prone to degradation in an aqueous environment and on disinfection. The survival capacity of these viruses depends on environmental temperature (Zheng et al., 2013). Behavior and survival of coronavirus in various aquatic environments have been discussed in Table 8.
Factors that can influence virus survival in water include temperature, organic matter, and aerobic microorganisms (Melnick et al., 1980; Sobsey & Meschke, 2003; John & Rose, 2005). The presence of protozoan and action of proteases and nucleases in water increase the inactivation rate of the viruses (Gerba et al., 1978; John & Rose, 2005).
Denaturation of a viral protein coat and cellular activity of viral enzymes decreases with increasing temperature (Hurst et al., 1989; John & Rose, 2005). Viruses are intercellular parasites and are active only within their hosts. In the case of COVID-19, it is not certain how long the virus survives on the surface. But its behavior resembles other coronaviruses. The survival of human coronaviruses on surfaces had large variability, ranging from 2 h to 9 days (Kampf et al., 2020). The enveloped viruses are less stable than non-enveloped viruses. Coronaviruses have a high degree of inactivation to temperature due to the enveloped coat. SARS-CoV identified in hospital wastewater, domestic sewage, and tap water were found to persist for 2 days at 20 °C and 14 days at 4 °C in wastewater. Coronaviruses are rapidly inactivated in wastewater with a 99.9% reduction in 2 to 3 days (Wang et al., 2005a).
The virus requires environmental vectors for their transmission (water, soil, metals, and hard surface). The many parameters including pH, salinity, temperature, and turbidity influence the persistence of viruses in the water (Melnick et al., 1980; Sobsey & Meschke, 2003; John & Rose, 2005). Viruses have high inactivation at greater temperature, i.e., at greater than 20 ℃ (John & Rose, 2005). Many experiments have been conducted to study the survival of coronavirus in water and wastewater. Coronavirus dies off rapidly in wastewater with a 99.9% reduction occurring in 2 to 3 days. The survival of the coronaviruses in primary wastewater is slightly longer than in secondary water due to the presence of a higher concentration of suspended solids which protects it from inactivation (Gundy et al., 2009).
To date, the COVID-19 virus survival in water or sewage is not evident due to its inactivation which is significantly rapid in comparison to non-enveloped human waterborne enteric viruses (adenovirus, norovirus, rotavirus etc.). Other human coronaviruses have survived for 2 days in dechlorinated tap water and hospital wastewater at 20 ℃ (Gundy et al., 2009). Inactivation of coronavirus is highly temperature-dependent and is affected by the concentration of organic matter and antagonistic bacteria in its environment. In tap water, coronaviruses are inactivated faster at 23 ℃ (10 days) than at 4 ℃. Coronaviruses die rapidly in wastewater, with T99.9 values between 2 and 4 days (Wang et al., 2005b).
Conclusion
The rate of survival of coronavirus in water or sewage is less. It is inactivated easily as present drinking water treatment plants disinfect viruses through filtration and other strategies. However, the presence of coronavirus or its genetic materials in wastewater will serve as a useful biomarker for wastewater based epidemiology and surveillance. Monitoring of the wastewater for the presence of pathogenic viruses will help in early warning and forecasting of disease outbreak and, hence, help in strategizing effective control measures.
References
Abdool Karim, S. S., & de Oliveira, T. (2021). New SARS-CoV-2 variants—clinical public health and vaccine implications. New England Journal of Medicine, 384(19), 1866–1868.
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O'Brien, J. W., & Tscharke, B. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Science of the Total Environment, 178, 138764.
Ahmed, W., Bertsch, P. M., Bivins, A., Bibby, K., Farkas, K., Gathercole, A., & Kitajima, M. (2020). Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Science of The Total Environment, 739, 139960.
Ali, S. A., Baloch, M., Ahmed, N., Ali, A. A., & Iqbal, A. (2020). The outbreak of coronavirus disease 2019 (COVID-19) - An emerging global health threat. Journal of Infection and Public Health, 13(4), 644–646.
Alygizakis, N., Markou, A. N., Rousis, N. I., Galani, A., Avgeris, M., Adamopoulos, P. G., Scorilas, A., Lianidou, E. S., Paraskevis, D., Tsiodras, S., Tsakris, A., Dimopoulos, M. A., & Thomaidis, N. S. (2021). Analytical methodologies for the detection of SARS-CoV-2 in wastewater: Protocols and future perspectives. Trends in Analytical Chemistry: TRAC, 134, 116125. https://doi.org/10.1016/j.trac.2020.116125
Asghar, H., Diop, O. M., Weldegebriel, G., Malik, F., Shetty, S., El Bassioni, L., & Burns, C. C. (2014). Environmental surveillance for polioviruses in the global polio eradication initiative. The Journal of Infectious Diseases, 210(suppl_1), S294-S303.
Ashbolt, N. J. (2004). Risk analysis of drinking water microbial contamination versus disinfection by-products (DBPs). Toxicology, 198(1–3), 255–262.
Bentham, R., & Whiley, H. (2018). Quantitative microbial risk assessment and opportunist waterborne infections–are there too many gaps to fill? International Journal of Environmental Research and Public Health, 15(6), 1150.
Bibby, K., Viau, E., & Peccia, J. (2011). Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Letters in Applied Microbiology, 52(4), 386–392.
Bibby, K., & Peccia, J. (2013). Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environmental Science & Technology, 47(4), 1945–1951.
Blanco, A., Abid, I., Al-Otaibi, N., Pérez-Rodríguez, F. J., Fuentes, C., Guix, S., & Bosch, A. (2019). Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food and Environmental Virology, 11(2), 184–192.
Carducci, A., Battistini, R., Rovini, E., & Verani, M. (2009). Viral removal by wastewater treatment: Monitoring of indicators and pathogens. Food and Environmental Virology, 1(2), 85–91.
Casanova, L., Rutala, W. A., Weber, D. J., & Sobsey, M. D. (2009). Survival of surrogate coronaviruses in water. Water Research, 43(7), 1893–1898.
Cavanagh, D. (2003). Severe acute respiratory syndrome vaccine development: Experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathology, 32(6), 567–582.
CDC, Centers for Disease control and Prevention. (2017). Guideline for isolation precautions: preventing transmission of infectious agents in healthcare settings. http://www.cdc.gov/hicpac/2007IP/2007isolationPrecautions. Accessed on 24th August 2020.
Choi, P. M., Tscharke, B., Samanipour, S., Hall, W. D., Gartner, C. E., Mueller, J. F., & O’Brien, J. W. (2019). Social demographic and economic correlates of food and chemical consumption measured by wastewater-based epidemiology. Proceedings of the National Academy of Sciences, 116(43), 21864–21873.
Colford, J. M., Jr., Roy, S., Beach, M. J., Hightower, A., Shaw, S. E., & Wade, T. J. (2006). A review of household drinking water intervention trials and an approach to the estimation of endemic waterborne gastroenteritis in the United States. Journal of Water and Health, 4(S2), 71–88.
Corman, V., Bleicker, T., Brünink, S., Drosten, C., & Zambon, M. (2020). Diagnostic detection of 2019-nCoV by real-time RT-PCR. World Health Organization, 17, 1–13.
Dada, A. C., & Gyawali, P. (2021). Quantitative microbial risk assessment (QMRA) of occupational exposure to SARS-CoV-2 in wastewater treatment plants. Science of The Total Environment, 763, 142989.
D'Aoust, P. M., Mercier, E., Montpetit, D., Jia, J. J., Alexandrov, I., Neault, N., Baig, A. T., Mayne, J., Zhang, X., Alain, T. and Langlois, M. A. (2021). Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water research, 188, 116560.
Daughton, C. G. (2001). Emerging pollutants and communicating the science of environmental chemistry and mass spectrometry: Pharmaceuticals in the environment. Journal of the American Society for Mass Spectrometry, 12(10), 1067–1076.
Daughton, C. G. (2018). Monitoring wastewater for assessing community health: Sewage chemical-information mining (SCIM). Science of the Total Environment, 619, 748–764.
Daughton, C. G. (2020). Wastewater surveillance for population-wide covid-19: The present and future. Science of the Total Environment, 736, 139631.
Dayaram, A., Franz, M., Schattschneider, A., Damiani, A. M., Bischofberger, S., Osterrieder, N., & Greenwood, A. D. (2017). Long term stability and infectivity of herpesviruses in water. Scientific Reports, 7, 46559.
Enteric viruses in drinking water document for public consultation prepared by the federal-provincial-territorial committee on drinking water. Accessed on May 28, 2020, from https://www.canada.ca/content/dam/hc-sc/documents/programs/consultation-enteric-virus-drinking-water/document/enteric-viruses-drinking-water.pdf
García-Aljaro, C., Blanch, A. R., Campos, C., Jofre, J., & Lucena, F. (2019). Pathogens faecal indicators and human-specific microbial source-tracking markers in sewage. Journal of Applied Microbiology, 126(3), 701–717.
Gerba, C. P., Stagg, C. H., & Abadie, M. G. (1978). Characterization of sewage solid-associated viruses and behavior in natural waters. Water Research, 12(10), 805–812.
Gonzalez-Reiche, A. S., Hernandez, M. M., Sullivan, M. J., Ciferri, B., Alshammary, H., Obla, A., & van Bakel, H. (2020). Introductions and early spread of SARS-CoV-2 in the New York City area. Science, 369(6501), 297–301.
Guan, Y., Zheng, B. J., He, Y. Q., Liu, X. L., Zhuang, Z. X., Cheung, C. L., & Butt, K. M. (2003). Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science, 302(5643), 276–278.
Gundy, P. M., Gerba, C. P., & Pepper, I. L. (2009). Survival of coronaviruses in water and wastewater. Food and Environmental Virology, 1(1), 10.
Haramoto, E., Malla, B., Thakali, O., & Kitajima, M. (2020). First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Science of The Total Environment, 737, 140405.
Hart, O.E., & Halden, R. U. (2020) Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility economy opportunities and challenges. Science of the Total Environment, 730, 138875.
Hasan, S. W., Ibrahim, Y., Daou, M., Kannout, H., Jan, N., Lopes, A., Alsafar, H., & Yousef, A. F. (2021). Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: Surveillance of COVID-19 epidemic in the United Arab Emirates. Science of The Total Environment, 764, 142929.
Heijnen, L., Elsinga, G., de Graaf, M., Molenkamp, R., Koopmans, M. P., & Medema, G. (2021). Droplet Digital RT-PCR to detect SARS-CoV-2 variants of concern in wastewater. medRxiv. https://doi.org/10.1101/2021.03.25.21254324
Hellmér, M., Paxéus, N., Magnius, L., Enache, L., Arnholm, B., Johansson, A., & Norder, H. (2014). Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Applied and Environmental Microbiology, 80(21), 6771–6781.
Hjelmso, M. H., Hellmer, M., Fernandez-Cassi, X., Timoneda, N., Lukjancenko, O., Seidel, M., & Abril, J. F. (2017). Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PloS one, 12(1), e0170199.
Hryniszyn, A., & Skonieczna, M. (2013). Methods for detection of viruses in water and wastewater. Advances in Microbiology, 3(05), 442.
Hurst, C. J., Benton, W. H., & Stetler, R. E. (1989). Detecting viruses in water. Journal-American Water Works Association, 81(9), 71–80.
Izda, V., Jeffries, M. A., & Sawalha, A. H. (2021). COVID-19: A review of therapeutic strategies and vaccine candidates. Clinical Immunology, 108634.
Izquierdo-Lara, R., Elsinga, G., Heijnen, L., Munnink, B. B. O., Schapendonk, C. M., Nieuwenhuijse, D., & De Graaf, M. (2021). Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. Emerging Infectious Diseases, 27(5), 1405.
John, D. E., & Rose, J. B. (2005). Review of factors affecting microbial survival in groundwater. Environmental Science & Technology, 39(19), 7345–7356.
Kampf, G., Todt, D., Pfaender, S., & Steinmann, E. (2020). Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. Journal of Hospital Infection, 104(3), 246–251.
Kasprzyk-Hordern, B., Bijlsma, L., Castiglioni, S., Covaci, A., de Voogt, P., Emke, E., & Thomas, K. V. (2014). Wastewater-based epidemiology for public health monitoring. Water and Sewerage Journal, 4, 25–26.
Kokkinos, P., Ziros, P., Meri, D., Filippidou, S., Kolla, S., Galanis, A., & Vantarakis, A. (2011). Environmental surveillance. An additional/alternative approach for virological surveillance in Greece?.International Journal of Environmental Research and Public Health, 8(6), 1914–1922.
Kumar, M., Patel, A. K., Shah, A. V., Raval, J., Rajpara, N., Joshi, M., & Joshi, C. G. (2020). First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Science of The Total Environment, 746, 141326.
La Rosa, G., Fratini, M., della Libera, S., Iaconelli, M., & Muscillo, M. (2012). Emerging and potentially emerging viruses in water environments. Annali Dell’istituto Superiore Di Sanità, 48, 397–406.
La Rosa, G., Bonadonna, L., Lucentini, L., Kenmoe, S., & Suffredini, E. (2020). Coronavirus in water environments: Occurrence persistence and concentration methods-A scoping review. Water Research, 179, 115899.
La Rosa, G., Mancini, P., Ferraro, G.B., Veneri, C., Iaconelli, M., Bonadonna, L., Lucentini, L., & Suffredini, E. (2021). SARS-CoV-2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Science of the total environment, 750, 141711.
Le Poder, S. (2011). Feline and canine coronaviruses: common genetic and pathobiological features. Advances in Virology, 2011.
Leung, W. K., To, K. F., Chan, P. K., Chan, H. L., Wu, A. K., Lee, N., & Sung, J. J. (2003). Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology, 125(4), 1011–1017.
Liu, X., Feng, J., Zhang, Q., Guo, D., Zhang, L., Suo, T., Hu, W., Guo, M., Wang, X., Huang, Z., & Xiong, Y. (2020). Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerging Microbes & Infections, 9(1), 1175–1179.
Mallapaty, S. (2020). How sewage could reveal true scale of coronavirus outbreak. Nature, 580(7802), 176–177.
Mao, K., Zhang, H., & Yang, Z. (2020). Can a paper-based device trace covid-19 sources with wastewater-based epidemiology? Environmental Science & Technology, 54, 3733–3735.
Martin, J., Klapsa, D., Wilton, T., Zambon, M., Bentley, E., Bujaki, E., Fritzsche, M., Mate, R., & Majumdar, M. (2020). Tracking SARS-CoV-2 in sewage: Evidence of changes in virus variant predominance during COVID-19 pandemic. Viruses, 12(10), 1144.
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., & Brouwer, A. (2020). Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environmental Science & Technology Letters, 7(7), 511–516.
Mellet, J., & Pepper, M. S. (2021). A covid-19 vaccine: Big strides come with big challenges. Vaccines (basel), 9(1), 39.
Melnick, J. L., Gerba, C. P., & Berg, G. (1980). The ecology of enteroviruses in natural waters. Critical Reviews in Environmental Science and Technology, 10(1), 65–93.
Miyani, B., Fonoll, X., Norton, J., Mehrotra, A., & Xagoraraki, I. (2020). SARS-CoV-2 in Detroit wastewater. Journal of Environmental Engineering, 146(11), 06020004.
Mlejnkova, H., Sovova, K., Vasickova, P., Ocenaskova, V., Jasikova, L., & Juranova, E. (2020). Preliminary study of SARS-CoV-2 occurrence in wastewater in the Czech Republic. International Journal of Environmental Research and Public Health, 17(15), 5508.
Nemudryi, A., Nemudraia, A., Wiegand, T., Surya, K., Buyukyoruk, M., Cicha, C., Vanderwood, K.K., Wilkinson, R. & Wiedenheft, B. (2020). Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Reports Medicine, 1(6), 100098.
Parasidis, T. A., Konstantinidis, T. G., & Alexandropoulou, I. G. (2013). Environmental monitoring of enteric viruses in wastewater. Virology & Mycology, 2, 1.
Peiris, J. S. M., Lai, S. T., Poon, L. L. M., Guan, Y., Yam, L. Y. C., Lim, W., & Cheng, V. C. C. (2003). Coronavirus as a possible cause of severe acute respiratory syndrome. The Lancet, 361(9366), 1319–1325.
Perez-Cataluna, A., Chiner-Oms, A., Cuevas Ferrando, E., Diaz-Reolid, A., Falco, I., Randazzo, W., & Sanchez Moragas, G. (2021). Detection Of Genomic Variants Of SARS-CoV-2 Circulating In Wastewater By High-Throughput Sequencing. medRxiv, 21251355.
Poon, L. L., Chan, K. H., Wong, O. K., Cheung, T. K., Ng, I., Zheng, B., & Peiris, J. S. (2004). Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clinical Chemistry, 50(1), 67–72.
Randazzo, W., Truchado, P., Cuevas-Ferrando, E., Simón, P., Allende, A., & Sánchez, G. (2020). SARS-CoV-2 RNA in wastewater anticipated covid-19 occurrence in a low prevalence area. Water research, 181, 115942.
Reslova, N., Michna, V., Kasny, M., Mikel, P., & Kralik, P. (2017). xMAP technology: applications in detection of pathogens. Frontiers in Microbiology, 8, 55.
Rimoldi, S.G., Stefani, F., Gigantiello, A., Polesello, S., Comandatore, F., Mileto, D., Maresca, M., Longobardi, C., Mancon, A., Romeri, F., & Pagani C. (2020). Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Science of the Total Environment, 744, 140911.
Saguti, F., Magnil, E., Enache, L., Churqui, M.P., Johansson, A., Lumley, D., Davidsson, F., Dotevall, L., Mattsson, A., Trybala, E., & Lagging, M. (2021). Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water research, 189, 116620.
Sims, N., & Kasprzyk-Hordern, B. (2020). Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environment International, 105689.
Sherchan S. P., Shahin S., Ward L. M., Tandukar S., Aw T. G., Schmitz B., Ahmed W., & Kitajima M. (2020). First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana USA. Science of the Total Environment, 743, 140621.
Sinclair, R. G., Jones, E. L., & Gerba, C. P. (2009). Viruses in recreational water-borne disease outbreaks: A review. Journal of Applied Microbiology, 107(6), 1769–1780.
Smeets, P. W. M. H., Rietveld, L. C., Van Dijk, J. C., & Medema, G. J. (2010). Practical applications of quantitative microbial risk assessment (QMRA) for water safety plans. Water Science and Technology, 61(6), 1561–1568.
Sobsey, M. D., & Meschke, J. S. (2003). Virus survival in the environment with special attention to survival in sewage droplets and other environmental media of fecal or respiratory origin. Report for the World Health Organization Geneva Switzerland, 70, 19–39.
Venugopal, A., Ganesan, H., Raja, S. S. S., Govindasamy, V., Arunachalam, M., Narayanasamy, A., & Vellingiri, B. (2020). Novel wastewater surveillance strategy for early detection of covid–19 hotspots. Current Opinion in Environmental Science & Health, 17, 8–13.
Wang, X. W., Li, J., Guo, T., Zhen, B., Kong, Q., Yi, B., Li, Z., Song, N., Jin, M., Xiao, W., & Zhu, X. (2005a). Waterborne pathogens-concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan hospital and the 309th Hospital of the Chinese People’s Liberation Army. Water Science and Technology, 52(8), 213–222.
Wang, X. W., Li, J. S., Guo, T. K., Zhen, B., Kong, Q. X., Yi, B., Li, Z., Song, N., Jin, M., Wu, X. M., & Xiao, W. J. (2005b). Excretion and detection of SARS coronavirus and its nucleic acid from digestive system. World Journal of Gastroenterology, 11(28), 4390.
Wang, X. W., Li, J. S., Jin, M., Zhen, B., Kong, Q. X., Song, N., Xiao, W. J., Yin, J., Wei, W., Wang, G. J., & Si, B. Y. (2005c). Study on the resistance of severe acute respiratory syndrome-associated coronavirus. Journal of Virological Methods, 126(1–2), 171–177.
Wang, P., Casner, R. G., Nair, M. S., Wang, M., Yu, J., Cerutti, G., & Ho, D. D. (2021). Increased resistance of SARS-CoV-2 variant P. 1 to antibody neutralization. Cell host & microbe, 29(5), 747–751.
Westhaus, A., Cabanes-Creus, M., Rybicki, A., Baltazar, G., Navarro, R. G., Zhu, E., & Lisowski, L. (2020). High-throughput in vitro, ex vivo, and in vivo screen of adeno-associated virus vectors based on physical and functional transduction. Human gene therapy, 31(9-10), 575–589.
White, M. K., Hu, W., & Khalili, K. (2015). The CRISPR/Cas9 genome editing methodology as a weapon against human viruses. Discovery Medicine, 19(105), 255.
Woolhouse, M. E., & Gowtage-Sequeria, S. (2005). Host range and emerging and reemerging pathogens. Emerging Infectious Diseases, 11(12), 1842.
World Health Organization. (2003). Guidelines for environmental surveillance of poliovirus circulation. http://polioeradication.org/wp-content/uploads/2016/07/WHO_V-B_03.03_eng.pdf.
World Health Organization. (2020). Naming the coronavirus disease (COVID-19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it
Wu, F., Zhang, J., Xiao, A., Gu, X., Lee, W.L., Armas, F., Kauffman, K., Hanage, W., Matus, M., Ghaeli, N. & Endo, N. (2020). SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. Msystems, 5(4).
Wurtzer, S., Marechal, V., Mouchel, J. M., Maday, Y., Teyssou, R., Richard, E., Almayrac, J. L., & Moulin, L. (2020). Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water Greater Paris France 5 March to 23 April 2020. Eurosurveillance, 25(50), 2000776.
Xagoraraki, I., & O’Brien, E. (2020). Wastewater-based epidemiology for early detection of viral outbreaks. In Women in Water Quality (pp. 75–97). Springer, Cham.
Ye, Y., Ellenberg, R. M., Graham, K. E., & Wigginton, K. R. (2016). Survivability partitioning and recovery of enveloped viruses in untreated municipal wastewater. Environmental Science & Technology, 50(10), 5077–5085.
Yuan, Y., Wang, N., & Ou, X. (2020). Caution should be exercised for the detection of SARS-CoV-2 especially in the elderly. Journal of Medical Virology, 92(9), 1641–1648.
Zaneti, R. N., Girardi, V., Spilki, F. R., Mena, K., Westphalen, A. P. C., da Costa Colares, E. R., & Etchepare, R. G. (2021). Quantitative microbial risk assessment of SARS-CoV-2 for workers in wastewater treatment plants. Science of The Total Environment, 754, 142163.
Zheng, C., Zhao, L., Zhou, X., Fu, Z., & Li, A. (2013). Treatment technologies for organic wastewater. Water Treatment, 11, 250–286.
Zhou, J. B., Kong, W. H., Wang, S., Long, Y. B., Dong, L. H., He, Z. Y., & Liu, M. Q. (2021). Detection of SARS-CoV-2 RNA in medical wastewater in Wuhan during the covid-19 outbreak. Virologica Sinica, 1.
Zuccato, E., Chiabrando, C., Castiglioni, S., Calamari, D., Bagnati, R., Schiarea, S., & Fanelli, R. (2005). Cocaine in surface waters: A new evidence-based tool to monitor community drug abuse. Environmental Health, 4(1), 14.
Author information
Authors and Affiliations
Contributions
The manuscript was conceptualized by Snehalatha Basavaraju, Jamuna Bai Aswathanarayan, Madhu Basavegowda, and Balasubramanian Somanathan. The original draft was prepared by Snehalatha Basavaraju and Jamuna Bai Aswathanarayan. The manuscript was reviewed and edited by Snehalatha Basavaraju, Jamuna Bai Aswathanarayan, Madhu Basavegowda, and Balasubramanian Somanathan. The manuscript was supervised by Balasubramanian Somanathan and Jamuna Bai Aswathanarayan.
Corresponding author
Ethics declarations
Consent for publication
All the authors have read and agreed for publishing the manuscript.
Competing interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Basavaraju, S., Aswathanarayan, J.B., Basavegowda, M. et al. Coronavirus: occurrence, surveillance, and persistence in wastewater. Environ Monit Assess 193, 508 (2021). https://doi.org/10.1007/s10661-021-09303-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-021-09303-8