Abstract
The SARS-CoV-2 spike (S) protein, a primary target for COVID-19 vaccine development, presents its receptor binding domain in two conformations, the receptor-accessible ‘up’ or receptor-inaccessible ‘down’ states. Here we report that the commonly used stabilized S ectodomain construct ‘2P’ is sensitive to cold temperatures, and this cold sensitivity is abrogated in a ‘down’ state-stabilized ectodomain. Our findings will impact structural, functional and vaccine studies that use the SARS-CoV-2 S ectodomain.
Similar content being viewed by others
Main
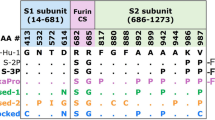
The spike (S) protein of SARS-CoV-2 mediates receptor binding and cell entry and is a key target for vaccine development efforts. Stabilized S ectodomain constructs have been developed that mimic the native spike, bind the ACE-2 receptor1,2 and present epitopes for neutralizing antibodies on their surface3,4,5,6. The so-called ‘2P’ S ectodomain construct (2P S) comprises residues 1–1,208 of SARS-CoV-2 S and contains two proline (2P) substitutions in the C-terminal S2 domain designed to stabilize the prefusion S conformation; a C-terminal foldon trimerization motif and a mutation that abrogates the furin-cleavage site1 (Fig. 1a). This and similar constructs have been widely used for structural biology and vaccine studies1,2,3,7,8. Purified S ectodomain proteins9 are assessed for quality control by SDS–PAGE, size exclusion chromatography (SEC), differential scanning fluorimetry (DSF)10 and negative-stain electron microscopy (NSEM). The last technique has been particularly informative because it reveals the structural integrity of individual molecules, allowing us to examine preparations that look similar by using bulk methods such as SDS–PAGE and SEC (Supplementary Fig. 1). The observed variability between preparations indicates a fragile S ectodomain, and measures to overcome the issue have been previously reported11,12.
a, Schematic of the SARS-CoV-2 spike (top, S) and a stabilized, furin-cleavage-deficient, soluble ectodomain construct (bottom, 2P S). b, Representative NSEM micrographs of 2P S samples: freshly prepared (left), after storage at 4 °C for 7 d (middle) or stored at 4 °C for 7 d followed by a 3-h incubation at 37 °C (right). Bar graph summarizes the results from NSEM analyses of 2P S samples stored under different conditions. Data shown are mean and range (for n = 2) or mean and s.e.m. (from n = 3–7) independent experiments with different protein lots; exact n at the top of each bar. c, Left, DSF profiles following changes in protein intrinsic fluorescence (expressed as a ratio between fluorescence at 350 and 330 nm) on applying a thermal ramp. Maxima and minima indicate inflection temperatures, Ti. For each storage condition (color coded as shown in d), five overlaid curves (technical replicates) are shown. Right, DSC profiles, shown as a representative curve of two technical replicates. d, Antibody binding to 2P S stored at different temperatures, measured by SPR; antibodies were CR3022 IgG (left) and 2G12 IgG (right). During the SPR run, the sample chamber was maintained at 37, 22 or 8 °C for samples that had been stored at 37, 22 or 4 °C, respectively. The binding experiments were carried out at 25 °C. Data are mean and s.e.m. of three technical replicates and are representative of at least five independent experiments, using separate protein lots (two independent repeats are shown in Extended Data Fig. 2).
Here we link the apparent fragility of 2P S to its rapid denaturation on storage at 4 °C (Fig. 1b). We followed the structural, biophysical and antigenic properties of 2P S stored under different temperature conditions (Fig. 1, Extended Data Figs. 1 and 2, Supplementary Tables 1 and 2 and Supplementary Figs. 2–6). 2P S was produced in 293F cells at 37 °C and purified at room temperature within 6–8 h (Supplementary Fig. 1). We performed NSEM analysis of 2P S incubated at different temperatures (Fig. 1b and Extended Data Fig. 1d,e). Freshly prepared 2P S samples assessed on the same day they were purified showed on average 75% well-formed spikes, with characteristic kite-shaped morphology on NSEM micrographs and two-dimensional (2D) class averages. This fraction slightly decreased to 64% after one cycle of freeze–thaw and to 59% after room temperature (22 °C) storage for 5–7 days, and it was substantially reduced to 5% after storage at 4 °C for 5–7 days. In contrast, after 1 week of storage at 37 °C, we observed 83% well-formed spikes. Furthermore, the well-formed spikes could be recovered after storage at 4 °C for 1 week with a 3-h incubation at 37 °C to roughly 75%, and three-dimensional (3D) classification of those particles showed the typical populations of the all-receptor-binding domain- (RBD) down and 1-RBD-up spike that we reported previously for freshly prepared samples9 (Supplementary Fig. 6). No further recovery from 4 °C storage was observed after longer incubations at 37 °C, which actually led to slight aggregation (Extended Data Fig. 1e).
SDS–PAGE analysis of 2P S samples stored at different temperatures did not indicate any appreciable protein degradation, although we did observe an increase in higher molecular weight bands for samples stored at 37 °C for 1 week (Supplementary Fig. 3). Similar to our NSEM results, we saw substantial differences in the quality and dispersion of particles in cryo-EM grids depending on the storage temperature of the specimen (Supplementary Fig. 4).
The S ectodomain was reported to exhibit conformational changes in response to pH12, so we asked whether the 2P S denaturation we observed was due to temperature-dependent pH changes in the Tris pH 8.0 buffer used13. A Tris buffer solution that measures pH 8.0 at 25 °C would measure roughly pH 8.6 at 4 °C. We performed SEC purification of a 2P S preparation into 3-morpholinopropane-1-sulfonic acid (MOPS) buffer pH 7.4; with smaller temperature dependence compared to Tris13, MOPS buffer should change only slightly, to pH 7.42, at 4 °C. Cold storage at pH 7.42 (MOPS buffer) reduced the spike fraction to 4%, similar to the average of 5% for cold storage in Tris buffer (Fig. 1b). Thus, temperature change appears to be the primary cause of 2P S denaturation, and not the pH shift within this range. We also tested the temperature effect at acidic pH and found that incubating 2P S at 4 °C in MES, pH 6 buffer reduced but did not eliminate the cold sensitivity.
Next, we examined whether the observed cold sensitivity is a property of the bulk protein in solution rather than an artifact of the NSEM sample preparation. First, we used a rapid DSF assay10 that measures changes in the protein intrinsic fluorescence as a thermal ramp is applied. The changes in fluorescence signal indicate transitions in the folding state of a protein, and the temperature at which a transition occurs is called the inflection temperature (Ti). Distinct profile shifts were observed for 2P S samples stored at different temperatures, with Ti shifted toward lower temperatures for samples stored at 4 °C, indicating lower protein stability, compared to samples stored at 22 or 37 °C (Fig. 1c and Supplementary Table 1). Next, we measured melting temperatures (Tm) using differential scanning calorimetry (DSC) (Fig. 1c, Supplementary Table 1 and Supplementary Fig. 5); after 1 week of storage at 37 °C, the 2P S sample featured an asymmetric unfolding transition with Tm of 65.5 °C. After 1 week of storage at 22 °C, we observed a second low-Tm transition at 48.2 °C. After 1 week of storage at 4 °C, we observed a similar two-peak profile with a markedly more pronounced low-Tm transition (Tm = 48.4 °C). On returning the 4 °C sample to 37 °C for 3 h before analysis, we observed an amplitude reduction of the low-Tm transition (Tm = 49.2 °C) and a corresponding amplitude increase in the high-Tm transition (Tm = 66.0 °C). Thus, the DSC results confirm that storage at 4 °C destabilizes 2P S compared to samples stored at 22 or 37 °C, and that returning the destabilized 2P S to 37 °C for 3 h substantially restores its stability, although the presence of the low-Tm peak suggests that the recovery is partial.
We next tested the effects of cold-induced instability on ligand binding to 2P S using surface plasmon resonance (SPR) and enzyme-linked immunosorbent assay (ELISA) (Fig. 1d and Extended Data Figs. 2 and 3). We found that 2P S stored at 4 °C showed higher binding to antibody CR3022 and to receptor ACE-2, which both require an ‘up’ RBD conformation for stable binding1,9,12, compared to freshly prepared protein. In contrast, cold storage reduced binding to antibody 2G12, which recognizes a quaternary glycan epitope in the S2 subunit14, indicating loss of quaternary structure during cold storage. We also tested two antibodies isolated from a COVID-19 convalescent donor, with epitopes mapped to the ACE-2 binding site (DH1179) and to the S2 region of spike (DH1189.1) (Extended Data Fig. 3 and Supplementary Figs. 7 and 8). Both antibodies showed different binding profiles depending on the temperature at which 2P S was stored. We next tested CR3022 and 2G12 binding to a 2P S sample that was flash frozen in liquid N2, stored in single-use aliquots at −80 °C and thawed by incubation at 37 °C (Supplementary Fig. 9). Freeze–thaw increased CR3022 binding to the 2P S compared to fresh sample, but incubating the thawed sample at 37 °C for 20 min reversed the effect, with binding activity similar to the levels observed with fresh sample.
These results indicated that cold-induced denaturation of the spike was associated with increased RBD-exposure of the S ectodomain. We thus asked whether a ‘down’ state-stabilized S ectodomain might be resistant to cold-induced denaturation (Fig. 2). Here we included a variant that combined the previously described rS2d9 and HexaPro mutations11 (Fig. 2a). This new variant, named rS2d-HexaPro, showed higher production yields compared to rS2d9. Similarly to rS2d9, rS2d-HexaPro particles appear 100% in a 3-RBD-down conformation as seen by NSEM (Fig. 2b, Supplementary Fig. 10 and Supplementary Video 1). Both HexaPro and rS2d-HexaPro appeared more resistant to cold-induced denaturation than 2P S (Fig. 1b), with rS2d-HexaPro showing a higher intact spike percentage than HexaPro (Fig. 2b). Thermostability and binding studies (Fig. 2c,d) further confirmed that HexaPro and rS2d-HexaPro were more resistant to cold destabilization compared to 2P S (Fig. 1).Two glycan-deleted mutants, 2P-N165A and 2P-N234A15, which have altered RBD-up propensity, showed substantial reduction of intact spike after incubation at 4 °C (Fig. 2f). The rS2d mutations (D985C, S383C) introduce an inter-protomer disulfide bond between the RBD and the S2 domain. Another study confirmed the stabilization effect of this disulfide bond, as well as that of an introduced disulfide bond between residues 987 and 413, for spikes stored at 4 °C (ref. 16). These data indicate that inter-promoter disulfide linkages may be a general strategy to stabilize the 2P S ectodomain and prevent its cold-induced denaturation.
a, Left, structures of HexaPro showing a 1-RBD-up conformation (PDB 6XKL) and rS2d (PDB 6X29) showing an all-RBD-down conformation. Middle, SDS–PAGE analysis of HexaPro and rS2d-HexaPro samples under reducing (R) or non-reducing (NR) conditions. Right, bar graph showing protein yields for rS2d and rS2d-HexaPro produced in 293F or CHO cells. b, Left, representative NSEM micrograph from a preparation of rS2d-HexaPro (top) and 2D class averages (bottom). Middle, 3D reconstruction of rS2d-HexaPro. Right, bar graph summarizing results from NSEM on S ectodomain variants stored at different temperatures. Data are mean and range (for n = 2) or s.e.m. (for n ≥ 3) for independent experiments with different protein lots. c, DSF profiles for HexaPro (left) and rS2d-HexaPro (right). For each storage condition, 3–4 overlaid curves (technical replicates) are shown. d, Binding of 2G12 and ACE-2 to HexaPro (left) and rS2d-HexaPro (right) measured by ELISA. OD450 nm, optical density at 450 nm.
Compared to 2P S, rS2d-HexaPro binding to ACE-2 was substantially reduced for samples stored at all temperatures, as expected for a spike fixed in an RBD-down conformation. Similarly, binding to 2G12 remained same for the rS2d-HexaPro construct irrespective of the storage temperature. For HexaPro, binding to ACE-2 was higher and 2G12 binding was reduced with 4 °C-stored protein compared to 37 °C-stored samples, showing that the HexaPro structure remained susceptible to perturbation at lower temperatures, whereas rS2d-HexaPro remains unperturbed on 4 °C storage.
Overall, our results demonstrate cold sensitivity of the furin-cleavage-deficient SARS-CoV-2 S ectodomain and highlight the importance of accounting for this effect, to obtain consistent results in serology assays or other binding studies using the 2P S or similar constructs. Based on these data, we recommend flash freezing spike ectodomain samples in single-use aliquots, then thawing at 37 °C followed by a brief (roughly 20 min) incubation at 37 °C before use in binding studies.
Methods
Protein expression and purification
SARS-CoV-2 ectodomain constructs were produced and purified as described previously1,11. Briefly, a gene encoding residues 1–1,208 of the SARS-CoV-2 S (GenBank MN908947) with proline substitutions at residues 986 and 987, a ‘GSAS’ substitution at the furin-cleavage site (residues 682–685), a C-terminal T4 fibritin trimerization motif, an HRV3C protease cleavage site, a TwinStrepTag and an 8×HisTag was synthesized and cloned into the mammalian expression vector pαH (Fig. 1a). Plasmids were transiently transfected into FreeStyle-293F cells using Turbo293 (SpeedBiosystems). Cells were grown in a 37 °C incubator with 9% CO2 and 120 r.p.m. rotary motion. Protein was purified on the sixth day post transfection from filtered supernatant using StrepTactin resin (IBA), followed by SEC purification using a Superose 6 10/300 Increase column in nCoV buffer (2 mM Tris, pH 8.0, 200 mM NaCl, 0.02% sodium azide). rS2d-HexaPro was also grown in CHO cells. Plasmids were transiently transfected into CHO cells using ExpiFectamine CHO Transfection Kit (Thermo Fisher), and protein was purified on the sixth day post transfection. All protein purification steps including Strep-tag purification and SEC were performed at room temperature. The spikes were purified the same day that the supernatant was harvested, and the purification was completed within 6–8 h. The purified protein was flash frozen and stored at −80 °C in single-use aliquots. Each aliquot was thawed and briefly incubated (roughly 20 min) at 37 °C before use.
Antibodies were produced in Expi293 cells and purified by Protein A affinity. For ACE-2 constructs, the ACE-2 C terminus was fused with either the human or mouse Fc region including a C-terminal 6× His-tag on the Fc domain. ACE-2 with human Fc tag was purified by Protein A affinity chromatography, and ACE-2 with mouse Fc tag was purified by Ni-NTA chromatography.
NSEM
Spike samples were incubated for the specified times in nCoV buffer at 4, 22 or 37 °C, then moved to room temperature to prepare NSEM grids, which was complete in less than 5 min. Samples were diluted to 100 µg ml−1 with room-temperature buffer containing 20 mM HEPES pH 7.4, 150 mM NaCl, 5% glycerol and 7.5 mM glutaraldehyde, and incubated for 5 min; then glutaraldehyde was quenched for 5 min by addition of 1 M Tris stock to a final concentration of 75 mM. A 5-µl drop of sample was applied to a glow-discharged, carbon-coated grid for 10–15 s, blotted, stained with 2% uranyl formate, blotted and air dried. Images were obtained with a Philips EM420 electron microscope at 120 kV, ×82,000 magnification and a 4.02 Å pixel size. The RELION program17 was used for particle picking, and 2D and 3D class averaging.
Thermostability assays
Thermostability of the S ectodomain samples was measured using DSF and DSC. Samples were purified and buffer exchanged into HEPES buffered saline (HBS) buffer (10 mM HEPES, 150 mM NaCl, pH 7.4) by SEC on a Superose 6 10/300 column. DSF assay was performed using Tycho NT. 6 (NanoTemper Technologies). Spike variants were diluted (0.15 mg ml−1) in HBS and run in triplicate. Intrinsic fluorescence was recorded at 330 and 350 nm while heating the sample from 35–95 °C at a rate of 30 °C min−1. The ratio of fluorescence (350/330 nm) and inflection temperatures (Ti) were calculated by Tycho NT. 6.
DSC measurements were performed using the NanoDSC platform (TA Instruments). Samples that had been incubated at 22 or 37 °C were purified by SEC at room temperature and the sample incubated at 4 °C was purified by SEC at 4 °C (Extended Data Fig. 2), diluted to 0.2–0.3 mg ml−1 in HBS, and degassed for 15 min at room temperature before analysis. DSC measurements were performed immediately after SEC purification of the samples. DSC cells were conditioned with filtered, degassed HBS before sample loading. Protein samples were heated from 10 °C to 100 °C at 1 °C min−1 under 3 atm pressure using HBS as the reference buffer. The observed denaturation profiles were buffer subtracted, converted to molar heat capacity, baseline-corrected with a sixth-order polynomial and fit with 2–4 Gaussian transition models, as needed, using the NanoAnalyze software (TA Instruments). The peak transition temperature (Tm) is reported as the temperature at the maximum observed heat capacity of each transition peak.
Isolation of antibodies from COVID-19 convalescent donors
Human SARS-CoV-2 spike antibodies DH1179 and DH1189.1 were isolated from a COVID-19 convalescent individual. Peripheral blood was collected following informed consent on a Duke University Medical Center approved Institutional Review Board protocol. Briefly, peripheral blood mononuclear cell samples collected after the onset of the symptoms were stained and the memory B cells were sorted with SARS-CoV-2 2P S probes. Antibody IgH and IgK/L genes were recovered from the single-cell sorted cells, cloned into human IgG1 constant region backbone and purified by Protein A beads as previously described18.
ELISA assays
Spike samples were preincubated at different temperatures then tested for antibody- or ACE-2-binding in ELISA assays as previously described19,20. Assays were run in two formats. In the first format, antibodies or ACE-2 protein were coated on 384-well plates at 2 µg ml−1 overnight at 4 °C, washed, blocked and followed by twofold serially diluted spike protein starting at 25 µg ml−1. Binding was detected with polyclonal anti-SARS-CoV-2 spike rabbit serum (developed in our laboratory), followed by goat anti-rabbit-HRP (Abcam no. ab97080) and 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Sera Care Life Sciences no. 5120-0083). Absorbance was read at 450 nm. In the second format, serially diluted spike protein was bound in individual wells of 384-well plates, which were previously coated with streptavidin (Thermo Fisher Scientific no. S-888) at 2 µg ml−1 and blocked. Proteins were incubated at room temperature for 1 h, washed, then human mAbs were added at 10 µg ml−1. Antibodies were incubated at room temperature for 1 h, washed and binding detected with goat anti-human-HRP (Jackson ImmunoResearch Laboratories, no. 109-035-098) and TMB substrate.
Commercially obtained constructs of SARS-CoV-2 spike ectodomain (S1+S2 ECD, S2 ECD and RBD) (Sino Biological Inc. catalog no. 40589-V08B1 and 40590-V08B, respectively, and RBD from Genescript catalog no. Z03483) were coated directly on 384-well plates at 2 µg ml−1 and incubated overnight at 4 °C. Plates were washed and blocked, and human mAbs threefold serially diluted from 100 µg ml−1 were added for 1 h at room temperature followed by washing. Binding was detected with goat anti-human IgG-HRP followed by TMB substrate.
Microneutralization assay
Live SARS-CoV-2 microneutralization assays were adapted from Berry et al.21 In short, sera or purified antibodies are diluted twofold and incubated with 100 median tissue culture infectious dose (TCID50) virus for 1 h. These dilutions are used as the input material for a TCID50. Each batch of microneutralization includes known neutralizing and non-neutralizing controls. Data are reported as the last concentration at which a test sample protects Vero E6 cells. Recently, a fluorescent infectious clone of SARS-CoV-2 was developed22.
Plaque reduction neutralization test (PRNT)
A live SARS-CoV-2 PRNT was performed in an assay adapted from Berry et al.21 Briefly, twofold dilutions of a test sample (serum, plasma, purified Ab) are incubated with 50 plaque forming units of SARS-CoV-2 for 1 h (NR-52281, BEI Resources). The antibody/virus mixture was then used to inoculate Vero E6 cells (ATCC). Cultures were then incubated at 37 °C, 5% CO2 for 1 h. At the end of the incubation, 1 ml of a viscous overlay (1:1 2× DMEM and 1.2% methylcellulose) was added to each well. Plates were incubated for 4 d. After fixation, staining and washing, plates were dried and plaques counted. Known neutralizing and non-neutralizing antibodies/sera are included in this assay as positive and negative controls, respectively. Plaques from each dilution of each sample were counted and data are reported as the concentration at which 50% of input virus was neutralized.
Pseudovirus neutralization assay
SARS-CoV-2 neutralization was assessed with Spike-pseudotyped viruses in 293T/ACE-2 cells as a function of reductions in luciferase (Luc) reporter activity. The 293T/ACE-2 cells were kindly provided by M. Farzan and H. Mu at Scripps. Cells were maintained in DMEM containing 10% FBS and 3 µg ml−1 of puromycin. An expression plasmid encoding codon-optimized full-length spike of the Wuhan-1 strain (VRC7480), was provided by B. Graham and K. Corbett at the Vaccine Research Center, National Institutes of Health. The D614G amino acid change was introduced into VRC7480 by site-directed mutagenesis using the QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent Technologies (catalog no. 210518). The mutation was confirmed by full-length spike gene sequencing. Pseudovirions were produced in human embryonic kidney 293T/17 cells (ATCC catalog no. CRL-11268) by transfection using Fugene 6 (Promega catalog no. E2692) and a combination of spike plasmid, lentiviral backbone plasmid (pCMV ΔR8.2) and firefly Luc reporter gene plasmid (pHR′ CMV Luc)23 in a 1:17:17 ratio. Transfections were allowed to proceed for 16–20 h at 37 °C. Medium was removed, monolayers rinsed with growth medium, and 15 ml of fresh growth medium added. Pseudovirus-containing culture medium was collected after an additional 2 d of incubation, and was clarified of cells by low-speed centrifugation and 0.45-µm micrometer filtration and stored in aliquots at −80 °C. TCID50 assays were performed on thawed aliquots to determine the infectious dose for neutralization assays (relative luminescence units (RLU) 500–1,000× background; the background usually averages 50–100 RLU).
For neutralization, a pretitrated dose of virus was incubated with eight serial three- or fivefold dilutions of serum samples or mAbs in duplicate in a total volume of 150 µl for 1 h at 37 °C in 96-well flat-bottom poly-l-lysine-coated culture plates (Corning Biocoat). Cells were suspended using TrypLE express enzyme solution (Thermo Fisher Scientific) and immediately added to all wells (10,000 cells in 100 µl of growth medium per well). One set of eight control wells received cells plus virus (virus control) and another set of eight wells received cells only (background control). After 66–72 h of incubation, medium was removed by gentle aspiration and 30 µl of Promega 1× lysis buffer was added to all wells. After a 10-min incubation at room temperature, 100 µl of Bright-Glo luciferase reagent was added to all wells. After 1–2 min, 110 µl of the cell lysate was transferred to a black/white plate (PerkinElmer). Luminescence was measured using a PerkinElmer Life Sciences, Model Victor2 luminometer. Neutralization titers are the serum dilution (inhibitory doses of 50 or 80%, ID50/ID80) or mAb concentration (inhibitory concentrations of 50 or 80%, IC50/IC80) at which RLU were reduced by 50 and 80% compared to virus control wells after subtraction of background RLU. Maximum percentage inhibition is the percentage neutralization at the lowest serum dilution or highest mAb concentration tested. Serum samples were heat-inactivated for 30 min at 56 °C before assay.
SPR
Antibody binding to SARS-CoV-2 spike constructs was assessed using SPR on a Biacore T-200 (GE Healthcare) with HBS buffer with 3 mM EDTA and 0.05% surfactant P-20 added. For the data presented in Fig. 1d and Extended Data Figs. 2 and 3, spike samples that were incubated at 4, 22 and 37 °C for 1 week were held in the sample chamber at 8, 22 and 37 °C, respectively, during the SPR run. The flow cell was maintained at 25 °C, thus all binding assays were performed at 25 °C. Antibodies captured on a CM5 chip coated with amine-coupled human Anti-Fc (8000RU) were assayed by SARS-CoV-2 spike at 200 nM. The surface was regenerated between injections with 3 M MgCl2 solution for 10 s at 100 µl min−1. Sensorgram data were analyzed using the BiaEvaluation software (GE Healthcare).
Cryo-EM
Purified SARS-CoV-2 spike preparations were diluted to a concentration of roughly 1 mg ml−1 in 2 mM Tris pH 8.0, 200 mM NaCl and 0.02% NaN3. A 2.5-µl drop of protein was deposited on a CF-1.2/1.3 grid that had been glow discharged for 30 s in a PELCO easiGlow Glow Discharge Cleaning System. After a 30 s incubation in >95% humidity, excess protein was blotted away for 2.5 s before being plunge frozen into liquid ethane using a Leica EM GP2 plunge freezer (Leica Microsystems). Frozen grids were imaged in a Titan Krios (Thermo Fisher) equipped with a K3 detector (Gatan).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The NSEM reconstruction for rS2d-HexaPro has been deposited in the Electron Microscopy Data Bank with accession code EMD-22934. NSEM reconstructions for the 2P spike after 1-week cold storage followed by 3-h recovery at 37 °C are deposited with accession codes EMD-22967 and EMD-22968 for the 3-RBD-down and 1-RBD-up states, respectively.
References
Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020).
Walls, A. C. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020).
Barnes, C. O. et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182, 828–842.e16 (2020).
Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020).
Mire, C. E. et al. A cross-reactive humanized monoclonal antibody targeting fusion glycoprotein function protects ferrets against lethal Nipah virus and hendra virus infection. J. Infect. Dis. 221, S471–S479 (2020).
Ke, Z. et al. Structures, conformations and distributions of SARS-CoV-2 spike protein trimers on intact virions. Nature 588, 498–502 (2020).
Liu, L. et al. Potent neutralizing monoclonal antibodies directed to multiple epitopes on the SARS-CoV-2 Spike. Nature 584, 450–456 (2020).
Zhou, T. et al. Structure-based design with tag-based purification and in-process biotinylation enable streamlined development of SARS-CoV-2 spike molecular probes. Cell Reports 33, https://doi.org/10.1016/j.celrep.2020.108322 (2020).
Henderson, R. et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 27, 925–933 (2020).
Magnusson, A. O. et al. nanoDSF as screening tool for enzyme libraries and biotechnology development. FEBS J. 286, 184–204 (2019).
Hsieh, L. et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020).
Zhou, T. et al. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains Cell Host Microbe https://doi.org/10.1016/j.chom.2020.11.004 (2020).
Good, N. E. et al. Hydrogen ion buffers for biological research. Biochemistry 5, 467–477 (1966).
Acharya, P. et al. A glycan cluster on the SARS-CoV-2 spike ectodomain is recognized by Fab-dimerized glycan-reactive antibodies. Preprint bioRxiv https://doi.org/10.1101/2020.06.30.178897 (2020).
Henderson, R. et al. Glycans on the SARS-CoV-2 spike control the receptor binding domain conformation. Preprint at bioRxiv https://doi.org/10.1101/2020.06.26.173765 (2020).
Xiong, X. et al. A thermostable, closed SARS-CoV-2 spike protein trimer. Nat. Struct. Mol. Biol. 27, 934–941 (2020).
Scheres, S. H. W. in Methods in Enzymology Vol. 579 (ed. Crowther, R. A.) 125–157 (Academic Press, 2016).
Liao, H. X. et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 (2013).
Alam, S. M. et al. Mimicry of an HIV broadly neutralizing antibody epitope with a synthetic glycopeptide. Sci. Transl. Med. 9, https://doi.org/10.1126/scitranslmed.aai7521 (2017).
Bonsignori, M. et al. Staged induction of HIV-1 glycan–dependent broadly neutralizing antibodies. Sci. Trans. Med. 9, https://doi.org/10.1126/scitranslmed.aai7514 (2017).
Berry, J. D. et al. Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus. J. Virol. Methods 120, 87–96 (2004).
Xie, X. et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27, 841–848.e3 (2020).
Naldini, L. et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267 (1996).
Acknowledgements
This work was supported by National Institutes of Health, National Institute of Allergy and Infectious Diseases extramural project grant nos. R01 AI145687 (P.A.) and AI058607 (G.D.S.), funding from the Department of Defense HR0011-17-2-0069 (G.D.S.) and a contract from the State of North Carolina Pandemic Recovery Office through funds from the Coronavirus Aid, Relief and Economic Security Act (B.F.H.). We thank J. McLellan for providing the 2P S plasmid. This work used the DSC platform supported by the Duke Consortia for HIV/AIDS Vaccine Development and the Titan Krios microscope in the Duke University Shared Materials Instrumentation Facility, a member of the North Carolina Research Triangle Nanotechnology Network, which is supported by the National Science Foundation (award no. ECCS-2025064) as part of the National Nanotechnology Coordinated Infrastructure. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281. Microneutralization and PRNT assays were performed in the Virology Unit of the Duke Regional Biocontainment Laboratory, which received partial support for construction from the NIH NIAID (grant no. UC6-AI058607).
Author information
Authors and Affiliations
Contributions
R.J.E., K. Mansouri., V.S. and P.A. discovered the effect of storage temperature on spike stability. R.J.E. led NSEM studies and established quantitative metrics for spike QC. K. Mansouri collected NSEM data and performed analyses. V.S. purified proteins and performed thermostability measurements. K. Manne purified proteins, performed SPR assays, prepared samples for NSEM, Tycho, ELISA, SPR and DSC measurements, and coordinated the study between the different research teams. B.W. performed DSC measurements. R.P., M.D., J.S. and W.W. performed ELISA assays. D.L., X.L., K.O.S. and G.D.S. isolated antibodies from convalescent patient. S.M.C.G. and K.J. purified proteins and performed thermostability assays. M.K purified proteins. Z.M. performed binding measurements. T.H.O., D.M. and G.D.S. performed neutralization assays. R.H. initiated the DSC experiments and provided the rS2d, 2P-N234A and 2P-N165A constructs before publication. S.M.A. supervised the DSC experiments. B.F.H. supervised ELISA experiments and antibody isolation. P.A. oversaw and led the study and cowrote the paper with R.J.E. and V.S. All authors reviewed and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Inês Chen was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 SDS-PAGE and NSEM analysis of SARS-CoV-2 2P S ectodomain incubated at different temperatures.
(a) SEC profile, (b) SDS-PAGE analysis (lane 1= molecular weight marker, lane 2= reducing conditions, lane 3= non-reducing conditions) and (c) DSF profile of a freshly purified sample of 2P S. (d) NSEM micrograph of 2P S after storage at 22 °C for one week. (e) NSEM micrograph of 2P S after storage at 4 °C for one week followed by recovery at 37 °C for 6 days. The circle indicates spike aggregation visible in the micrograph.
Extended Data Fig. 2 Changes in antigenicity of SARS-CoV-2 2PS ectodomain incubated at different temperatures.
(a) Antibody CR3022 IgG (left) and 2G12 IgG (right) binding to two independently purified lots of 2P spike stored at different temperatures measured by SPR. Data for spike samples measured after a 1-week incubation at 37, 22, and 4 °C, are shown in blue, green, and red respectively; sample stored 1 week at 4 °C and then incubated for 6 hours at 37 °C shown in cyan. During the SPR run the sample chamber was maintained at temperatures of 37 °C, 22 °C and 8 °C, for the 37 °C, 22 °C and 4 °C incubated samples, respectively. The binding experiments were carried out at 25 °C. The bar graphs in Fig. 1d show data from Lot # 025MFK. The schematics shows the assay format. (b) ELISA binding profiles showing binding of ACE-2 receptor ectodomain, RBD-directed antibody CR3022, and S2-directed, glycan-reactive antibody, 2G12. The same color scheme was used as for the SPR experiment. The black line indicates a freshly purified 2P spike sample that was flash frozen, then thawed and incubated for 20 min at 37 °C. The schematics show the ELISA format. The measurements in the top row were done in a format where antibody was coated on the plate and the measurements in the bottom row were done in a format where spike was captured on a strep-coated plate (see Methods).
Extended Data Fig. 3 Antigenic response of SARS-CoV-2 2P S ectodomain incubated at different temperatures to antibodies elicited from convalescent patient sera.
(a) SPR binding profiles showing binding of (left) RBD-directed antibody, DH1179 and (right) S2-directed antibody, DH1189.1 to spike samples incubated for 1 week at either 37 °C (blue), 22 °C (green) or 4 °C (red). Binding to spike sample first incubated at 4 °C for 1 week, then moved to 37 °C for 3 hours prior to the experiment is shown in cyan. (b) ELISA binding profiles showing binding of RBD-directed antibody, DH1179, S2-directed antibody, DH1189.1, and influenza HA-directed antibody CH65 (control) to spike samples incubated at different temperatures (same color scheme as in panel a). The black line indicates a freshly purified 2P S sample that was flash-frozen, then thawed and incubated for 20 min at 37 °C.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Figs. 1–10.
Supplementary Video 1
Structural variability in RBD and N-terminal domain (NTD) dispositions in rS2d-HexaPro revealed by 3D classification of NSEM data.
Rights and permissions
About this article
Cite this article
Edwards, R.J., Mansouri, K., Stalls, V. et al. Cold sensitivity of the SARS-CoV-2 spike ectodomain. Nat Struct Mol Biol 28, 128–131 (2021). https://doi.org/10.1038/s41594-020-00547-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-00547-5
This article is cited by
-
Structural and biochemical rationale for Beta variant protein booster vaccine broad cross-neutralization of SARS-CoV-2
Scientific Reports (2024)
-
Impact of SARS-CoV-2 spike stability and RBD exposure on antigenicity and immunogenicity
Scientific Reports (2024)
-
Influence of variant-specific mutations, temperature and pH on conformations of a large set of SARS-CoV-2 spike trimer vaccine antigen candidates
Scientific Reports (2023)
-
Stability and expression of SARS-CoV-2 spike-protein mutations
Molecular and Cellular Biochemistry (2023)
-
Structure-based design of stabilized recombinant influenza neuraminidase tetramers
Nature Communications (2022)