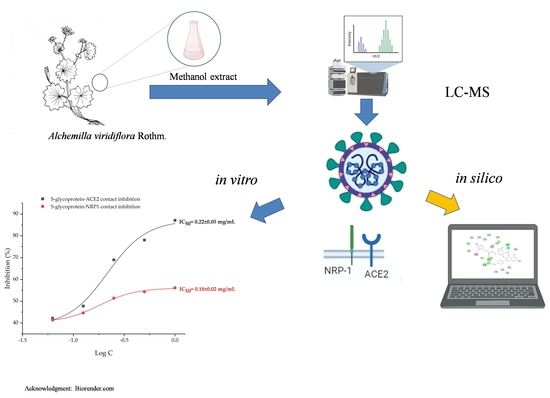
In Silico and In Vitro Studies of Alchemilla viridiflora Rothm—Polyphenols’ Potential for Inhibition of SARS-CoV-2 Internalization
Abstract
:1. Introduction
2. Results and Discussion
2.1. LC-MS Chemical Analysis (Phytochemical Analysis)
2.2. Molecular Docking Studies
2.3. In Vitro SARS-CoV-2 Internalization Inhibition Assays
3. Materials and Methods
3.1. Plant Material and Extract Preparation
3.2. Chemicals
3.3. LC-MS Chemical Analysis
3.4. Molecular Docking Simulations
3.4.1. Dataset
3.4.2. Docking Parameters
3.5. Molecular Dynamics (MD) Simulation
3.6. In Vitro SARS-CoV-2 Internalization Inhibition Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pohlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Muller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Harwansh, R.K.; Bahadur, S. Herbal Medicines to Fight Against COVID-19: New Battle with an Old Weapon. Curr. Pharm. Biotechnol. 2022, 23, 235–260. [Google Scholar] [CrossRef]
- Živković, J.; Suručić, R.; Arsenijević, J. Beneficial Effects of Polyphenolics from Fruit Species in Prevention and Management of Type 2 Diabetes. In A Closer Look at Polyphenolics; Bertollini, P., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2022. [Google Scholar]
- Grabez, M.; Skrbic, R.; Stojiljkovic, M.P.; Vucic, V.; Rudic Grujic, V.; Jakovljevic, V.; Djuric, D.M.; Surucic, R.; Savikin, K.; Bigovic, D.; et al. A prospective, randomized, double-blind, placebo-controlled trial of polyphenols on the outcomes of inflammatory factors and oxidative stress in patients with type 2 diabetes mellitus. Rev. Cardiovasc. Med. 2022, 23, 57. [Google Scholar] [CrossRef] [PubMed]
- Goc, A.; Sumera, W.; Rath, M.; Niedzwiecki, A. Phenolic compounds disrupt spike-mediated receptor-binding and entry of SARS-CoV-2 pseudo-virions. PLoS ONE 2021, 16, e0253489. [Google Scholar] [CrossRef] [PubMed]
- Xiu, S.; Dick, A.; Ju, H.; Mirzaie, S.; Abdi, F.; Cocklin, S.; Zhan, P.; Liu, X. Inhibitors of SARS-CoV-2 Entry: Current and Future Opportunities. J. Med. Chem. 2020, 63, 12256–12274. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.J.; Huynh, T.K.; Yang, C.S.; Hu, D.W.; Shen, Y.C.; Tu, C.Y.; Wu, Y.C.; Tang, C.H.; Huang, W.C.; Chen, Y.; et al. Hesperidin Is a Potential Inhibitor against SARS-CoV-2 Infection. Nutrients 2021, 13, 2800. [Google Scholar] [CrossRef]
- Tito, A.; Colantuono, A.; Pirone, L.; Pedone, E.; Intartaglia, D.; Giamundo, G.; Conte, I.; Vitaglione, P.; Apone, F. Pomegranate Peel Extract as an Inhibitor of SARS-CoV-2 Spike Binding to Human ACE2 Receptor (in vitro): A Promising Source of Novel Antiviral Drugs. Front. Chem. 2021, 9, 638187. [Google Scholar] [CrossRef] [PubMed]
- Surucic, R.; Tubic, B.; Stojiljkovic, M.P.; Djuric, D.M.; Travar, M.; Grabez, M.; Savikin, K.; Skrbic, R. Computational study of pomegranate peel extract polyphenols as potential inhibitors of SARS-CoV-2 virus internalization. Mol. Cell Biochem. 2021, 476, 1179–1193. [Google Scholar] [CrossRef] [PubMed]
- Surucic, R.; Travar, M.; Petkovic, M.; Tubic, B.; Stojiljkovic, M.P.; Grabez, M.; Savikin, K.; Zdunic, G.; Skrbic, R. Pomegranate peel extract polyphenols attenuate the SARS-CoV-2 S-glycoprotein binding ability to ACE2 Receptor: In silico and in vitro studies. Bioorg. Chem. 2021, 114, 105145. [Google Scholar] [CrossRef]
- Makau, J. Anti-influenza activity of Alchemilla mollis extract: Possible virucidal activity against influenza virus particles. Drug Discov. Ther. 2013, 7, 189–195. [Google Scholar]
- Filippova, E.I. Antiviral Activity of Lady’s Mantle (Alchemilla vulgaris L.) Extracts against Orthopoxviruses. Bull. Exp. Biol Med. 2017, 163, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Radovic, J.; Surucic, R.; Niketic, M.; Kundakovic-Vasovic, T. Alchemilla viridiflora Rothm.: The potent natural inhibitor of angiotensin I-converting enzyme. Mol. Cell Biochem. 2022, 477, 1893–1903. [Google Scholar] [CrossRef]
- Hippisley-Cox, J.; Young, D.; Coupland, C.; Channon, K.M.; Tan, P.S.; Harrison, D.A.; Rowan, K.; Aveyard, P.; Pavord, I.D.; Watkinson, P.J. Risk of severe COVID-19 disease with ACE inhibitors and angiotensin receptor blockers: Cohort study including 8.3 million people. Heart 2020, 106, 1503–1511. [Google Scholar] [CrossRef] [PubMed]
- Silva Fernandes, A.; Hollanda Veras, J.; Silva, L.S.; Puga, S.C.; Luiz Cardoso Bailao, E.F.; de Oliveira, M.G.; Cardoso, C.G.; Carneiro, C.C.; Costa Santos, S.D.; Chen-Chen, L. Pedunculagin isolated from Plinia cauliflora seeds exhibits genotoxic, antigenotoxic and cytotoxic effects in bacteria and human lymphocytes. J. Toxicol. Environ. Health A 2022, 85, 353–363. [Google Scholar] [CrossRef]
- Tamura, S.; Yang, G.M.; Yasueda, N.; Matsuura, Y.; Komoda, Y.; Murakami, N. Tellimagrandin I, HCV invasion inhibitor from Rosae Rugosae Flos. Bioorg. Med. Chem. Lett. 2010, 20, 1598–1600. [Google Scholar] [CrossRef]
- Tian, J.; Xie, Y.; Zhao, Y.; Li, C.; Zhao, S. Spectroscopy characterization of the interaction between brevifolin carboxylic acid and bovine serum albumin. Luminescence 2011, 26, 296–304. [Google Scholar] [CrossRef]
- Zhan, Y.; Ta, W.; Tang, W.; Hua, R.; Wang, J.; Wang, C.; Lu, W. Potential antiviral activity of isorhamnetin against SARS-CoV-2 spike pseudotyped virus in vitro. Drug Dev. Res. 2021, 82, 1124–1130. [Google Scholar] [CrossRef]
- Fakih, T.M.; Dewi, M.L. In silico Identification of Characteristics Spike Glycoprotein of SARS-CoV-2 in the Development Novel Candidates for COVID-19 Infectious Diseases. J. Biomed. Transl. Res. 2020, 6, 48–52. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. MedRxiv 2020. [Google Scholar] [CrossRef]
- Available online: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 (accessed on 27 January 2022).
- Shuster, A.; Pechalrieu, D.; Jackson, C.B.; Abegg, D.; Choe, H.; Adibekian, A. Clinical Antiviral Drug Arbidol Inhibits Infection by SARS-CoV-2 and Variants through Direct Binding to the Spike Protein. ACS Chem. Biol. 2021, 16, 2845–2851. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.H.; Lee, J.; Jeon, S.; Kim, S.; Min, J.S.; Kwon, S. Natural Polyphenols, 1,2,3,4,6-O-Pentagalloyglucose and Proanthocyanidins, as Broad-Spectrum Anticoronaviral Inhibitors Targeting Mpro and RdRp of SARS-CoV-2. Biomedicines 2022, 10, 1170. [Google Scholar] [CrossRef] [PubMed]
- Yasmin, T.; Ali, M.T.; Haque, S.; Hossain, M. Interaction of Quercetin of Onion with Axon Guidance Protein Receptor, NRP-1 Plays Important Role in Cancer Treatment: An In Silico Approach. Interdiscip. Sci. 2017, 9, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Perez-Miller, S.; Patek, M.; Moutal, A.; Cabel, C.R.; Thorne, C.A.; Campos, S.K.; Khanna, R. In silico identification and validation of inhibitors of the interaction between neuropilin receptor 1 and SARS-CoV-2 Spike protein. bioRxiv 2020. [Google Scholar] [CrossRef]
- Parker, M.W.; Xu, P.; Li, X.; Vander Kooi, C.W. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 2012, 287, 11082–11089. [Google Scholar] [CrossRef] [PubMed]
- Fischer, U.A.; Carle, R.; Kammerer, D.R. Identification and quantification of phenolic compounds from pomegranate (Punica granatum L.) peel, mesocarp, aril and differently produced juices by HPLC-DAD-ESI/MS(n). Food Chem. 2011, 127, 807–821. [Google Scholar] [CrossRef]
- Liu, X.; Raghuvanshi, R.; Ceylan, F.D.; Bolling, B.W. Quercetin and Its Metabolites Inhibit Recombinant Human Angiotensin-Converting Enzyme 2 (ACE2) Activity. J. Agric. Food Chem. 2020, 68, 13982–13989. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Anton-Plagaro, C.; Shoemark, D.K.; Simon-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, A.; Allerston, C.K.; Jia, H.; Herzog, B.; Garza-Garcia, A.; Winfield, N.; Ellard, K.; Aqil, R.; Lynch, R.; Chapman, C.; et al. Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction. J. Med. Chem. 2010, 53, 2215–2226. [Google Scholar] [CrossRef] [PubMed]
- Guedes, I.A.; Costa, L.S.C.; Dos Santos, K.B.; Karl, A.L.M.; Rocha, G.K.; Teixeira, I.M.; Galheigo, M.M.; Medeiros, V.; Krempser, E.; Custodio, F.L.; et al. Drug design and repurposing with DockThor-VS web server focusing on SARS-CoV-2 therapeutic targets and their non-synonym variants. Sci. Rep. 2021, 11, 5543. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Dunbrack, R.L., Jr.; Hooft, R.W.; Krieger, B. Assignment of protonation states in proteins and ligands: Combining pKa prediction with hydrogen bonding network optimization. Methods Mol. Biol. 2012, 819, 405–421. [Google Scholar] [PubMed]






| Compound | Formula: | Molecular Weight: | Match Score: | RT: | Adduct/Loss: |
|---|---|---|---|---|---|
| Pedunculagin | C34H24O22 | 784.076 | 0.998 | 9.35 | −/H+ |
| Galloyl-HHDP hexose | C27H22O17 | 618.086 | 0.999 | 12.41 | Na+/− |
| Isoquercitrin | C21H20O12 | 464.095 | 0.999 | 12.44 | H+/− |
| Quercetin 3-(6″-ferulylglucoside) | C31H28O15 | 640.143 | 0.993 | 12.46 | −/H+ |
| Tellimagrandin I | C34H26O22 | 786.092 | 0.993 | 16.4 | −/H+ |
| Brevifolin carboxylic acid | C13H8O8 | 292.022 | 0.997 | 21.5 | −/H2OH+ |
| Myricetin 3-O-glucuronide | C21H18O14 | 494.07 | 0.973 | 22.9 | CH3OHH+/− |
| Tellimagrandin II | C41H30O26 | 938.103 | 0.992 | 23.97 | −/H+ |
| Pentagalloylglucose | C41H32O26 | 940.118 | 0.879 | 29.36 | −/H+ |
| Kaempferol 7-O-glucuronide | C21H18O12 | 462.08 | 0.996 | 30.97 | Na+/− |
| HHDP-hexoside | C20H18O14 | 482.07 | 0.961 | 31.1 | CH3OHH+/− |
| Quercetin 3-methyl ether 7-glucuronide | C22H20O13 | 492.09 | 0.985 | 31.13 | −/H+ |
| Kaempferol 7-O-glucoside | C21H20O11 | 448.101 | 0.981 | 33.06 | Na+/− |
| Di-O-methylquercetin | C17H14O7 | 330.074 | 0.999 | 33.82 | −/H+ |
| Tiliroside | C30H26O13 | 594.137 | 0.996 | 37.7 | −/H+ |
| Isorhamnetin-3-O-glucoside | C22H22O12 | 478.111 | 0.963 | 39.37 | NH4+/− |
| Miquelianin | C21H18O13 | 478.075 | 0.96 | 39.37 | NH4+/− |
| Compound | Bind Energy [kcal/mol] | Interacting Residues * |
|---|---|---|
| Quercetin 3-(6″-ferulylglucoside) | −8.035 | Gln160, Glu151 (1.63 Å), Phe157 (2.63 Å), Ser161 (1.84 Å), Tyr 162 (2.83 Å) |
| Tellimagrandin I | −8.022 | Gln160 (2.82 Å), Glu151 (1.57 Å, 1.71 Å), Phe157 (2.82 Å, 2.84 Å) |
| Tellimagrandin II | −7.955 | Gln160, Glu151 (1.59 Å, 1.64 Å), Gly163 (3.10 Å), Tyr116, Tyr116 (1.73 Å, 1.94 Å), Tyr162 (2.24 Å) |
| Pedunculagin | −7.848 | Gln160 (2.23 Å), Glu151 (1.64 Å, 2.15 Å), Gly163 (2.70 Å), Leu119, Phe157, Tyr116 (1.69 Å), Tyr162 (2.46 Å) |
| Isorhamnetin-3-O-glucoside | −7.761 | Glu151 (1.63 Å, 1.76 Å), Leu119, Leu159 (1.60 Å), Phe157, Ser161 (2.92 Å), Tyr162 (2.79 Å) |
| Tiliroside | −7.633 | Arg70, Gln73 (1.50 Å), Gln160, Glu151 (1.61 Å), Lys84 (2.73 Å), Phe157 (2.42 Å), Tyr120 |
| Pentagalloylglucose | −7.601 | Gln160 (2.52 Å), Gln160, Glu151 (1.54 Å, 1.62 Å), Ser161 (2.64 Å), Tyr156, Tyr162 (2.02 Å) |
| Kaempferol 7-O-glucuronide | −7.519 | Glu151 (1.92 Å), Phe157 (2.43 Å), Tyr120 (1.20 Å) |
| Di-O-methylquercetin | −7.515 | Gln160 (2.38 Å), Glu151 (1.59 Å, 1.60 Å), Phe123, Phe157 (2.33 Å), Tyr156 |
| HHDP-hexoside | −7.506 | Glu151 (1.71 Å, 2.00 Å), Phe157, Ser161 (2.39 Å), Tyr116 (2.34 Å) |
| Miquelianin | −7.406 | Gln160 (3.09 Å), Glu151 (1.72 Å, 1.92 Å), Leu119, Phe157, Ser161 (1.49 Å) |
| Myricetin 3-O-glucuronide | −7.404 | Glu151 (1.67 Å, 1.94 Å), Leu119, Leu159 (1.73 Å), Phe157, Ser161 (1.46 Å) |
| Umifenovir ** | −7.384 | Glu151 (1.65 Å), Ser161 (1.96 Å), Tyr116 |
| Quercetin ** | −7.189 | Gln160 (2.07 Å), Glu151 (1.63 Å, 1.78 Å), Phe123, Phe157 (2.02 Å), Tyr156 |
| Kaempferol 7-O-glucoside | −7.121 | Gln160 (2.48 Å, 3.02 Å), Glu151 (1.66 Å, 1.70 Å), Phe157 (1.97 Å), Tyr162 (1.90 Å) |
| Galloyl-HHDP hexose | −6.964 | Gln160 (2.56 Å, 2.77 Å), Glu151 (1.56 Å, 1.82 Å), Leu159 (1.70 Å, 1.96 Å) |
| Isoquercitrin | −6.953 | Gln160 (3.03 Å), Glu151 (1.63 Å, 1.64 Å), Leu159 (1.89 Å), Ser161 (1.80 Å, 2.32 Å) |
| Quercetin 3-methyl ether 7-glucuronide | −6.579 | Glu151 (1.58 Å, 1.62 Å), Lys84 (3.06 Å), Lys84, Tyr120 (1.62 Å) |
| Brevifolin carboxylic acid | −6.359 | Arg70 (1.53 Å), Tyr162 (1.84 Å), Tyr172 (1.63 Å) |
| Compound | The Most Favorable Binding Pose ** | Bind Energy [kcal/mol] | Interacting Residues * |
|---|---|---|---|
| Pentagalloylglucose |  | −7.685 | Asp320 (2.29 Å, 2.50 Å), Lys351 (2.29, 2.32 Å), Lys351, Lys352 (2.04 Å), Pro317 (2.50 Å), Thr413 (2.61 Å, 2.68 Å), Tyr297, Tyr353 (2.41 Å, 2.71 Å) |
| Quercetin methyl ether glucuronide |  | −7.667 | Asp320 (2.72 Å), Glu348 (2.09 Å), Lys351 (2.07 Å), Thr413, Thr413 (3.01 Å), Trp301 (2.00 Å), Trp411,Tyr297, Tyr353 (2.51 Å) |
| Tiliroside |  | −7.594 | Asp320, Glu348 (2.15 Å), Lys351 (2.05 Å), Thr413, Tyr297, Tyr353 (2.82 Å) |
| Kaempferol 7-O-glucuronide |  | −7.452 | Asn300 (1.82 Å), Asp320, Lys351 (2.05 Å), Trp301 (2.36 Å), Tyr297 |
| Kaempferol 7-O-glucoside |  | −7.264 | Asn300 (1.85 Å), Asp320, Lys351 (2.02 Å), Trp301 (2.30 Å), Tyr297 |
| Quercetin |  | −7.205 | Asn300 (2.60 Å), Asp320, Lys351 (1.94 Å, 2.85 Å), Thr349 (2.72 Å), Trp301 (2.28 Å), Tyr297 (2.51 Å), Tyr353 |
| Miquelianin |  | −6.986 | Asp320 (2.44 Å), Glu348 (2.65 Å), Lys351 (1.98 Å), Thr349 (2.60 Å), Thr413 |
| Brevifolin carboxylic acid *** |  | −6.976 | Lys351 (2.00 Å), Thr316, Trp301 (2.32 Å), Trp301, Tyr297 |
| Quercetin 3-(6″-ferulylglucoside) |  | −6.875 | Arg418 (2.65 Å), Arg418, Asn309 (2.90 Å), Asn313 (2.37 Å),Glu312, Ile345, Lys350 (2,84 Å), Ser346 (2.83 Å), Thr388 |
| Pedunculagin |  | −6.855 | Asn300 (2.78 Å), Asp320, Ser298, Tyr297 (2.33 Å, 2.49 Å) |
| Myricetin 3-O-glucuronide |  | −6.802 | Asp320, Gly318 (2.17 Å), Lys351 (1.99 Å), Thr349 (2.72 Å),Thr413, Tyr297 |
| Isoquercitrin |  | −6.799 | Asp320, Gly318 (2.52 Å), Lys351 (2.03 Å, 2.52 Å), Ser346 (3.02 Å), Thr413, Tyr297 |
| Tellimagrandin I |  | −6.76 | Asp320, Glu319 (2.77 Å), Gly318, Ser321, Thr413 (1.93 Å), Tyr297 |
| Di-O-methylquercetin |  | −6.713 | Asn300 (2.05 Å), Glu348, Tyr297, Tyr301, Tyr353 |
| Isorhamnetin-3-O-glucoside |  | −6.467 | Asp320, Asp320 (2.71 Å), Gly318, Lys351 (2.25 Å, 2.40 Å), Ser346 (2.65 Å), Thr413, Thr413 (2.56 Å), Tyr297 |
| Galloyl-HHDP-hexose |  | −6.365 | Asp320, Asp320 (2.07 Å, 2.92 Å), Thr413, Trp301, Trp411 (2.51 Å), Tyr297, Tyr353 (2.67 Å) |
| Tellimagrandin II |  | −6.051 | Arg323, Asp320 (2.76 Å), Tyr297 |
| HHDP-hexoside |  | −5.897 | Lys351 (2.02 Å), Thr413 (2.46 Å), Tyr353 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suručić, R.; Radović Selgrad, J.; Kundaković-Vasović, T.; Lazović, B.; Travar, M.; Suručić, L.; Škrbić, R. In Silico and In Vitro Studies of Alchemilla viridiflora Rothm—Polyphenols’ Potential for Inhibition of SARS-CoV-2 Internalization. Molecules 2022, 27, 5174. https://doi.org/10.3390/molecules27165174
Suručić R, Radović Selgrad J, Kundaković-Vasović T, Lazović B, Travar M, Suručić L, Škrbić R. In Silico and In Vitro Studies of Alchemilla viridiflora Rothm—Polyphenols’ Potential for Inhibition of SARS-CoV-2 Internalization. Molecules. 2022; 27(16):5174. https://doi.org/10.3390/molecules27165174
Chicago/Turabian StyleSuručić, Relja, Jelena Radović Selgrad, Tatjana Kundaković-Vasović, Biljana Lazović, Maja Travar, Ljiljana Suručić, and Ranko Škrbić. 2022. "In Silico and In Vitro Studies of Alchemilla viridiflora Rothm—Polyphenols’ Potential for Inhibition of SARS-CoV-2 Internalization" Molecules 27, no. 16: 5174. https://doi.org/10.3390/molecules27165174