Abstract
The novel coronavirus disease 2019 (COVID-19) presents with non-specific clinical features. This may result in misdiagnosis or delayed diagnosis, and lead to further transmission in the community. We aimed to derive early predictors to differentiate COVID-19 from influenza and dengue. The study comprised 126 patients with COVID-19, 171 with influenza and 180 with dengue, who presented within 5 days of symptom onset. All cases were confirmed by reverse transcriptase polymerase chain reaction tests. We used logistic regression models to identify demographics, clinical characteristics and laboratory markers in classifying COVID-19 versus influenza, and COVID-19 versus dengue. The performance of each model was evaluated using receiver operating characteristic (ROC) curves. Shortness of breath was the strongest predictor in the models for differentiating between COVID-19 and influenza, followed by diarrhoea. Higher lymphocyte count was predictive of COVID-19 versus influenza and versus dengue. In the model for differentiating between COVID-19 and dengue, patients with cough and higher platelet count were at increased odds of COVID-19, while headache, joint pain, skin rash and vomiting/nausea were indicative of dengue. The cross-validated area under the ROC curve for all four models was above 0.85. Clinical features and simple laboratory markers for differentiating COVID-19 from influenza and dengue are identified in this study which can be used by primary care physicians in resource limited settings to determine if further investigations or referrals would be required.
Similar content being viewed by others
Introduction
A cluster of cases of pneumonia with unknown cause was detected in Wuhan in December 20191. Caused by a novel human coronavirus now named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), it has since spread rapidly on a global scale. On 11 March 2020, the World Health Organization (WHO) declared the novel coronavirus disease (COVID-19) as a pandemic, and expressed deep concern about the alarming levels of spread and severity2. Confirmation of acute SARS-CoV-2 infection requires detection of the virus in respiratory samples by reverse transcriptase polymerase chain reaction (RT-PCR) test. Clinical presentation of COVID-19 patients ranges from asymptomatic to mild non-specific acute symptoms such as fever, dry cough and fatigue3. Close to 20% of COVID-19 cases may be severe4.
Influenza is prevalent globally and remains an important cause of morbidity and mortality from respiratory viral infections5 while dengue is prevalent in tropical countries with geographic expansion6. Dengue is also known as not only the leading cause of fever in returning travelers in non-endemic countries, but also the main source for triggering autochthonous transmissions7,8. Since clinical presentations of these common viral infections are non-specific, it is difficult for primary care physicians to differentiate COVID-19 from influenza and dengue. This may result in misdiagnosis or delayed diagnosis, and lead to further transmission in the community. As the COVID-19 pandemic progresses, doctors in both dengue endemic9,10,11 and non-endemic countries where influenza may be the most relevant differential diagnosis for COVID-195,12, should maintain a high level of suspicion and be provided with effective tools to differentiate these three infections.
Singapore is a tropical city state with several endemic viral infections. Influenza circulates year round with bimodal peaks typically observed in April-July and November-January13. Dengue epidemics of increasing magnitude have occurred in a five- to six-year cycle affecting more adults than children14. As of 15 June 2020, a total of 40,818 cases of the COVID-19 including 26 deaths have been reported in Singapore15. It is of concern that two local cases of COVID-19 with false positive dengue serology were reported in Singapore without travel or contact history16.
In this study, we compared clinical presentations and laboratory parameters of patients with COVID-19, influenza and dengue. We constructed predictive models using logistic regression with the aim to assist doctors in differentiating COVID-19 from influenza and dengue based on clinical features and simple laboratory investigations to determine if further investigations or referrals would be required.
Methods
Cohort description
Our study population comprised laboratory confirmed COVID-19 from an ongoing prospective cohort, and influenza and dengue patients from previous studies. Briefly, prospective recruitment of COVID-19 patients took place from January to April 2020 at the National Centre for Infectious Diseases (NCID), which is a purpose-built facility designed to strengthen Singapore’s capabilities in infectious disease management3. All COVID-19 suspect cases, i.e., clinical signs and symptoms suggestive of pneumonia or severe respiratory infection with breathlessness or acute respiratory illness of any severity, were sent to NCID and other hospitals via dedicated ambulances for testing and isolation during the study period.
Influenza patients were selected from a retrospective study of those who presented at Tan Tock Seng Hospital (TTSH), the primary centre for screening, treatment and isolation of influenza A(H1N1)pdm09 cases from April to June 2009 in Singapore17. Dengue patients were selected from a prospective cohort study where adult patients presented with acute undifferentiated febrile illness to TTSH from January 2010 to September 201218.
Data collection and confirmatory testing
At first presentation to the hospital, demographic data, symptoms and signs were collected. Full blood count, renal and liver function tests were performed. SARS-CoV-2, influenza and dengue viruses were detected in respiratory samples or venous blood by RT-PCR tests3,17,18. Data of 215 patients with COVID-19 (all with wild-type strain), 200 with H1N1(pdm09) or seasonal influenza, and 300 with dengue (predominantly dengue virus serotype 2) were extracted. As our objective was to derive early predictors to differentiate COVID-19 from influenza and dengue, we analysed only symptomatic patients who presented to hospital within 5 days after symptom onset.
Ethics consideration
The study procedures were carried out in accordance with relevant guidelines and regulations. Ministry of Health (MOH) and the National Healthcare Group Domain Specific Review Board (NHG DSRB), Singapore approved the studies (A Multi-centred Prospective Study to Detect Novel Pathogens and Characterize Emerging Infections (E/12/917), Prospective adult dengue study (E/09/432) and Characterization of influenza in local setting including novel influenza A 2009 cases (E/09/344)). Data for COVID-19 patients were collected with waiver of informed consent granted by MOH under the Infectious Diseases Act as part of COVID-19 outbreak investigation. Research samples and data for the prospective COVID-19 and dengue cohort studies were collected with written informed consent approved by NHG DSRB. Data from influenza patients were collected with waiver of informed consent approved by NHG DSRB as stated earlier.
Statistical analysis
Fisher’s exact test was used to compare categorical variables and Mann–Whitney U test to compare continuous variables between any two groups. All statistical tests were two-sided, and statistical significance was taken as P < 0.05.
We used multivariable logistic regression models as predictive tools to differentiate between COVID-19 and influenza, and COVID-19 and dengue. We followed the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) 2015 guideline19.
Two multivariable models were fitted for differentiating COVID-19 versus influenza (flu models 1 and 2) and COVID-19 versus dengue (dengue models 1 and 2): flu model 1 and dengue model 1 contained demographics and symptoms, whereas flu model 2 and dengue model 2 included laboratory parameters in addition to demographics and symptoms.
For the analysis on COVID-19 versus influenza, the proportion of missing data for laboratory parameters ranged from 3.4% (10 out of total 297 observations) for WBC count, haemoglobin, platelet, neutrophil count and lymphocyte count, to 28.3% (84 observations) for albumin, alanine aminotransferase (ALT) and aspartate aminotransferase (AST). The variables for flu models 1 and 2 were selected through stepwise use of Akaike’s information criterion. Multicollinearity was then checked using variance inflation factor (VIF) for the final predictor variables included in these 2 multivariable models, which ranged from 1.01 to 1.25.
For the analysis on COVID-19 versus dengue, the proportion of missing data for laboratory parameters ranged from 3.3% (10 out of total 306 observations) for WBC count, haemoglobin, haematocrit, platelet, neutrophil count and lymphocyte count, to 9.2% (28 observations) for AST. As the data for differentiating COVID-19 versus dengue exhibited separation, finite estimates of adjusted odds ratios (aOR) and Wald confidence intervals (CI) could not be obtained. Hence, we applied Firth’s modified score procedure to estimate OR and derived CI using the profile-penalized likelihood function20. The variables in dengue models 1 and 2 were selected by backward stepwise approach using penalized likelihood ratio tests. The VIF for the final predictor variables ranged from 1.01 to 1.23 for dengue model 1, and it ranged from 1.31 to 3.67 in dengue model 2 when laboratory parameters were included.
We assessed the predictive performance of the multivariable logistic regression models using receiver operating characteristic (ROC) curves and the corresponding area under the ROC (AUC). As the evaluation of the predictive performance of a set of independent variables using the original sample may yield an overly optimistic estimate of how the results would generalize to an independent set of data, we applied a tenfold cross-validation to obtain a more realistic estimate of AUC for each model.
Results
This study comprised 126 patients with COVID-19, 171 with influenza and 180 with dengue, who presented within 5 days after symptom onset. The demographic characteristics of the patients are shown in Table 1. The median age of COVID-19 patients was older compared with influenza and dengue patients. A lower proportion of COVID-19 patients were male compared with dengue patients. The proportion of COVID-19 patients having comorbidities was higher compared with dengue patients but not with influenza patients.
The clinical features of patients with COVID-19, influenza and dengue at presentation are shown in Table 2. Shortness of breath and diarrhoea were more common in COVID-19 patients than in influenza patients, while fever, cough, running nose and sore throat were less common. Cough, shortness of breath, running nose and sore throat were more common in COVID-19 patients than in dengue patients. A lower proportion of COVID-19 patients had fever, diarrhoea, muscle aches, fatigue/malaise, abdominal pain, bleeding, conjunctivitis, headache, joint pain, skin rash and vomiting/nausea compared with dengue patients. We also provided an infographic of percentage of COVID-19, influenza and dengue patients with each symptom at presentation. It can be seen that COVID-19 and influenza patients have similar symptoms, while dengue patients present with symptoms that are significantly different from the other two groups (See Supplementary Table S1).
The vital signs and laboratory parameters of patients with COVID-19, influenza and dengue are shown in Table 3. COVID-19 patients had lower white blood cell (WBC) count, neutrophil count and creatinine compared with influenza patients whereas their lymphocyte count and alanine aminotransferase (ALT) were higher. WBC, platelet, neutrophil and lymphocyte counts and albumin were higher in COVID-19 patients than dengue patients. The levels of haemoglobin, haematocrit, aspartate aminotransferase (AST) and creatinine were lower in COVID-19 patients than dengue patients.
The multivariable logistic regressions differentiating COVID-19 from influenza are shown in Fig. 1. In flu model 1 containing demographics and symptoms, older age (aOR 1.09; 95% CI 1.07–1.12), shortness of breath (aOR 18.29; 95% CI 2.28–411.81) and diarrhoea (aOR 13.70; 95% CI 2.33–128.89) increased the odds that the patient had COVID-19, while fever, cough, running nose and vomiting/nauseas were indicative of influenza. In flu model 2 containing demographics, symptoms and laboratory parameters, older age (aOR 1.10; 95% CI 1.07–1.13), shortness of breath (aOR 50.66; 95% CI 3.09–1391.20), diarrhoea ((aOR 8.59; 95% CI 1.67–67.62) and higher lymphocyte count (aOR 1.93; 95% CI 1.09–3.46) were predictive of COVID-19, while cough, running nose and lower neutrophil count were indicative of influenza.
Final covariates in multivariable logistic regression for differentiating COVID-19 versus influenza: (A) model 1 on demographics and symptoms, (B) model 2 on demographics, symptoms and laboratory Adjusted odds ratios (aOR) and 95% confidence interval (CI)l for having COVID-19 are shown on log10 scale.
Figure 2 shows the multivariable logistic regression analysis differentiating COVID-19 versus dengue. In dengue model 1 containing demographics and symptoms, older age (aOR 1.06; 95% CI 1.01–1.12) increased the odds that the patient had COVID-19, while fever, headache, joint pain, skin rash, vomiting/nauseas and bleeding were indicative of dengue. In dengue model 2 containing demographics, symptoms and laboratory parameters, patients who had cough (aOR 51.48; 95% CI 4.47–4,662.18), higher platelet count (aOR 1.04; 95% CI 1.01–1.09) and higher lymphocyte count (aOR 213.28; 95% CI 9.65–98,867.53) were at increased odds of COVID-19, while cough, headache, joint pain, skin rash and vomiting/nauseas were indicative of dengue.
Final covariates in multivariable logistic regression for differentiating COVID-19 versus dengue: (A) model 1 on demographics and symptoms, (B) model 2 on demographics, symptoms and laboratory parameters. Adjusted odds ratios (aOR) and 95% confidence interval (CI) for having COVID-19 are shown on log10 scale.
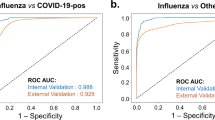
The cross-validated AUC of flu model 1 containing demographics and symptoms was 0.871 (95% CI 0.829–0.914), and the cross-validated AUC of flu model 2 which included laboratory parameters in differentiating COVID-19 versus influenza was 0.890 (95% CI 0.851–0.929) (Fig. 3). The cross-validated AUC of dengue model 1 without laboratory parameters and of dengue model 2 which included laboratory parameters for differentiating COVID-19 versus dengue were 0.995 (95% CI 0.989–1.000) and 0.997 (95% CI 0.993–1.000) respectively (Fig. 4).
Discussion
Early identification of COVID-19 suspected cases is critical for effective surveillance and successful containment of disease spread. Cases who are not diagnosed correctly and in a timely manner may lead to further transmission of the virus21. It is imperative to make available a rapid, sensitive, and affordable point-of-care screening test in the primary care setting for early detection of COVID-1922. There is possible cross-reactivity between dengue virus and SARS-CoV-2 antibodies, which can lead to false-positive dengue serology among COVID-19 patients and vice versa because of possible similarities between SARS-CoV-2 epitopes in the HR2-domain of the spike-protein and the dengue envelope-protein23. In September 2020, the WHO published an interim guidance on the use of SARS-CoV-2 antigen-detecting rapid diagnostic tests (Ag-RDTs) in countries or areas that are experiencing widespread community transmission, where the health system may be over-burdened, and where it may not be possible to test all or any suspect cases by RT-PCR tests. It is recommended to use the Ag-RDT for the suspected symptomatic cases unless the person is a contact of a confirmed COVID-19 case24. To ensure that suspected cases are detected as early as possible, primary care physicians who serve as the first point of contact in the healthcare system need a reliable predictive tool to differentiate COVID-19 from other viral infections such as influenza and dengue at first presentation.
In this study, we built logistic regression models using clinical features and simple laboratory investigations to differentiate COVID-19 from influenza and dengue (Figs. 1 and 2). In view of the lower sensitivity of flu model 1 when only symptoms were considered for differentiation of COVID-19 from influenza (Fig. 3), it is important to consider full blood count for further assessment where appropriate. Our study revealed that shortness of breath was the most important symptom for differentiating COVID-19 from influenza. Patients with cough and higher platelet count were more likely to have COVID-19, while headache, joint pain, skin rash and vomiting/nausea were indicative of dengue. These clinical features and simple laboratory parameters can serve as a guide for preliminary screening especially for primary care physicians in Singapore and other tropical countries where dengue is endemic and influenza virus circulates all-year round.
Decision algorithms for differentiating dengue, influenza and other febrile illnesses have been reported before25. There were a few studies on prediction models for diagnosis of COVID-19, however, these included measurement of cytokines, computed tomography scan or genome sequencing26. Sun et al. had come up with algorithms for estimating the risk of COVID-19 among patients who presented to NCID for SARS-CoV-2 testing, which included a subset of the same cohort of COVID-19 cases in our study, but the duration of symptom onset was not taken into consideration and controls included all SARS-CoV-2 negative patients regardless of final diagnosis27. In the study by Sun et al., the AUC of the logistic regression model that predicted COVID-19 was 0.65 when only demographics and clinical variables were considered, whereas the models with laboratory and exposure-risk variables included had higher AUCs of 0.88 to 0.9127.
As our study relied on simple parameters to differentiate COVID-19 from influenza and dengue, the predictors identified from the multivariable logistic regression models may guide primary care physicians, who serve as the first point of contact with the healthcare system, in deciding who should be tested with RT-PCR tests or Ag-RDTs especially in the absence of travel history to high-risk countries and epidemiological links16. In countries and remote regions where confirmatory diagnostic capabilities are limited28, clinical features and simple laboratory parameters identified in this study may be used as one of the criteria for isolation of suspected cases to prevent further transmission arising from prolonged delay from symptom onset to isolation29.
There are a few limitations in our study. We included patients who presented to hospital within 5 days of symptom onset, hence the findings may not be applicable to those presented late for medical care. Patients who were not suspected or sufficiently ill to be referred to hospitals were excluded. The patients in our study were recruited in three different periods as it was not feasible to recruit influenza and dengue patients during the COVID-19 outbreak in 2020; (i) there was a drastic decline in influenza activity and (ii) non-COVID-19 related research activities were put on hold to minimize the risk of COVID-19 transmission. There may be differences in clinical features and laboratory parameters at presentation due to the predominant strains of SARS-CoV-2 virus among COVID-19 patients, virus serotypes among dengue patients, and virus strains among influenza patients circulating at different time points. Few COVID-19 cases co-infected with dengue had been reported thus far in Singapore, Thailand and French island of Mayotte30,31,32. Case series of pneumonia patients co-infected with COVID-19 and influenza were also reported in China33. In Singapore, after the implementation of public health measures for COVID-19, a marked decline in influenza activity had been reported34. There is an urgent need for clinical, epidemiological and laboratory studies to be undertaken to better understand the interactions between COVID-19, influenza and dengue.
In conclusion, we have shown that multivariable models based on clinical features and simple laboratory markers for differentiating COVID-19 from influenza and dengue, which possess good predictive performance can serve as a useful tool for primary care physicians to determine if further investigations or referrals would be required. The findings from our study would need to be further validated, so as to address the knowledge gaps of the ongoing COVID-19 pandemic.
Data availability
The datasets analysed during the current study are available from the corresponding author on reasonable request.
References
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
World Health Organization. Coronavirus disease 2019 (COVID-19) Situation Report 52. (2020). Available at: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200312-sitrep-52-covid-19.pdf?sfvrsn=e2bfc9c0_4. (Accessed: 27th May 2020).
Young, B. E. et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 323, 1488–1494 (2020).
Wu, Z. & McGoogan, J. M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA 323, 1239–1242 (2020).
Xu, X. et al. Update: Influenza activity in the United States during the 2018–19 season and composition of the 2019–20 influenza vaccine. MMWR Morb. Mortal. Wkly. Rep. 68, 544–551 (2019).
Stanaway, J. D. et al. The global burden of dengue: An analysis from the Global Burden of Disease Study 2013. Lancet Infect. Dis. 16, 712–723 (2016).
Wilder-Smith, A., Ooi, E.-E., Horstick, O. & Wills, B. Dengue. Lancet 393, 350–363 (2019).
Monge, S. et al. Characterization of the first autochthonous dengue outbreak in Spain (August-September 2018). Acta Trop. 205, 105402 (2020).
Young, B. E. & Chen, M. Influenza in temperate and tropical Asia: A review of epidemiology and vaccinology. Hum. Vaccin. Immunother. 16, 1659–1667 (2020).
Torres, M. C. et al. Re-introduction of dengue virus serotype 2 in the state of Rio de Janeiro after almost a decade of epidemiological silence. PLoS ONE 14, e0225879 (2019).
Navarro, J.-C., Arrivillaga-Henríquez, J., Salazar-Loor, J. & Rodriguez-Morales, A. J. COVID-19 and dengue, co-epidemics in Ecuador and other countries in Latin America: Pushing strained health care systems over the edge. Travel Med. Infect. Dis. https://doi.org/10.1016/j.tmaid.2020.101656 (2020).
Bordi, L. et al. Differential diagnosis of illness in patients under investigation for the novel coronavirus (SARS-CoV-2), Italy, February 2020. Euro Surveill. 25, 2000170 (2020).
Ministry of Health, Singapore. Communicable Diseases Surveillance in Singapore 2018. (2019).
Rajarethinam, J. et al. Dengue in Singapore from 2004 to 2016: Cyclical epidemic patterns dominated by serotypes 1 and 2. Am. J. Trop. Med. Hyg. 99, 204–210 (2018).
Ministry of Health, Singapore. Updates on COVID-19 (Coronavirus Disease 2019) Local Situation. (2020). Available at: https://www.moh.gov.sg/covid-19 (Accessed: 16th June 2020).
Yan, G. et al. Covert COVID-19 and false-positive dengue serology in Singapore. Lancet. Infect. Dis 20, 536 (2020).
Ong, A. K. et al. Improving the clinical diagnosis of influenza–a comparative analysis of new influenza A (H1N1) cases. PLoS ONE 4, e8453 (2009).
Leo, Y.-S. et al. Utility of warning signs in guiding admission and predicting severe disease in adult dengue. BMC Infect. Dis. 13, 498 (2013).
Moons, K. G. M. et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): Explanation and elaboration. Ann. Intern. Med. 162, W1-73 (2015).
Heinze, G. A comparative investigation of methods for logistic regression with separated or nearly separated data. Stat. Med. 25, 4216–4226 (2006).
Munster, V. J., Koopmans, M., van Doremalen, N., van Riel, D. & de Wit, E. A novel coronavirus emerging in China—key questions for impact assessment. N. Engl. J. Med. 382, 692–694 (2020).
Wong, J. E. L., Leo, Y. S. & Tan, C. C. COVID-19 in Singapore—current experience: Critical global issues that require attention and action. JAMA 323, 1243–1244 (2020).
Lustig, Y. et al. Potential antigenic cross-reactivity between SARS-CoV-2 and Dengue viruses. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciaa1207 (2020).
World Health Organization. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays, Interim guidance, 11 September 2020. https://www.who.int/publications-detail-redirect/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays (2020). (Accessed: 30th September 2020)
Low, J. G. H. et al. The early clinical features of dengue in adults: challenges for early clinical diagnosis. PLoS Negl. Trop. Dis. 5, e1191 (2011).
Wynants, L. et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ 369, 1328 (2020).
Sun, Y. et al. Epidemiological and clinical predictors of COVID-19. Clin. Infect. Dis. 71, 786–792 (2020).
Gilbert, M. et al. Preparedness and vulnerability of African countries against importations of COVID-19: A modelling study. Lancet 395, 871–877 (2020).
Hellewell, J. et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob. Health 8, e488–e496 (2020).
Teo, J. & Zhou, T. Coronavirus: Rare to have Covid-19 and dengue, say experts of Singapore’s first patient with both. The Straits Times (2020). Available at: https://www.straitstimes.com/singapore/health/rare-to-have-coronavirus-and-dengue (Accessed: 31st March 2020)
Tun, S. Thailand records first coronavirus death: health official. Reuters 2, 2 (2020).
Epelboin, L., Blondé, R., Nacher, M., Combe, P. & Collet, L. COVID-19 and dengue co-infection in a returning traveller. J. Travel Med. https://doi.org/10.1093/jtm/taaa114 (2020).
Ding, Q., Lu, P., Fan, Y., Xia, Y. & Liu, M. The clinical characteristics of pneumonia patients coinfected with novel coronavirus and influenza virus in Wuhan, China. J. Med. Virol. 2, 2. https://doi.org/10.1002/jmv.25781 (2020).
Soo, R. J. J., Chiew, C. J., Ma, S., Pung, R. & Lee, V. Decreased Influenza Incidence under COVID-19 Control Measures, Singapore. Emerg. Infect. Dis. 26, 1933–1935 (2020).
Acknowledgements
This study was supported by National Medical Research Council, Singapore.
Author information
Authors and Affiliations
Contributions
Y.S.L. and D.C.L. designed the study. T.L.T. and L.W.A. reviewed literature. T.L.T., B.Y. and M.I.C. extracted the data. L.W.A. analysed the data. T.L.T., L.W.A., M.I.C., Y.S.L. and D.C.L. interpreted the data. T.L.T. and L.W.A. wrote the drafts of the manuscript. B.Y., M.I.C., Y.S.L. and D.C.B.L. commented on and helped revise the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
Dr. Young reports personal fees from Sanofi and Roche, outside the submitted work. All other authors have no potential competing interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Thein, TL., Ang, L.W., Young, B.E. et al. Differentiating coronavirus disease 2019 (COVID-19) from influenza and dengue. Sci Rep 11, 19713 (2021). https://doi.org/10.1038/s41598-021-99027-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99027-z
This article is cited by
-
A review on the significance of body temperature interpretation for early infectious disease diagnosis
Artificial Intelligence Review (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.