Detection of SARS-CoV-2 Genome in Stool and Plasma Samples of Laboratory Confirmed Iranian COVID-19 Patients
- 1Basic and Molecular Epidemiology of Gastrointestinal Disorders Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Microbiology and Microbial Biotechnology, Faculty of Life Sciences and Biotechnology, Shahid Beheshti University, Tehran, Iran
- 3Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 4Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 5Department of Anesthesiology, Faculty of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 6Research and Development Center, Imam Hossein Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Coronavirus disease 2019 (COVID19), caused by the severe acute respiratory syndrome coronavirus 2 (SARSCoV2), was first discovered in China in late 2019 and quickly spread worldwide. Although nasopharyngeal swab sampling is still the most popular approach identify SARS-CoV-2 carriers, other body samples may reveal the virus genome, indicating the potential for virus transmission via non-respiratory samples. In this study, researchers looked at the presence and degree of SARS-CoV-2 genome in stool and plasma samples from 191 Iranian COVID-19 patients, and looked for a link between these results and the severity of their disease. SARS-CoV-2 RNA shedding in feces and plasma of COVID-19 patients was assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Medical data were collected and evaluated, including Clinical features, demographics, radiological, and laboratory findings of the patients. Plasma samples from 117 confirmed laboratory patients were evaluated and 24 out of 117 patients (20.51%) tested positive for SARS-COV-2 RNA. Besides, 20 out of 74 patients (27.03%) tested positive for SARS-COV-2 RNA in stool samples. There seems to be no relationship between the presence of SARS-CoV-2 genome in fecal and plasma samples of Covid-19 patients and the severity of illness. We provide evidence of the SARS-CoV-2 genome presence in stool and plasma samples of Iranian COVID-19 patients.
1 Introduction
In late December 2019, a series of strange pneumonia cases were reported in China, with symptoms that were strikingly similar to viral pneumonia, and soon spread to other parts of the globe (Zhu et al., 2020). The Chinese Center for Disease Control and Prevention (CDC) found a novel coronavirus in a patient’s throat sample in early January 2020, and the World Health Organization (WHO) named it SARS-COV-2 (Cheung et al., 2020). Before SARS-CoV-2, two other outbreaks were reported by two other members of the coronavirus family, occurred in 2002 by SARS-CoV and in 2012 by MERS-CoV, respectively (Lippi et al., 2020). The SARS-CoV-2, which belongs to the lineage B of the genus Beta coronaviruses, has a single-stranded RNA-positive genome with about 30,000 nucleotides length and is very similar to the SARS-CoV (Wu et al., 2020). But unlike the previous two outbreaks, SARS-CoV-2 has caused millions of deaths in terms of severe respiratory complications, mainly dyspnea and other organ failures (Lippi et al., 2020). In addition to the usual symptoms of respiratory diseases, other symptoms such as chest pain, headache, and especially gastrointestinal symptoms are also observed in patients. The most common clinical indications of SARS-COV-2 infection are vague, and many of these symptoms may also be caused by other viruses that cause respiratory tract infections (Udugama et al., 2020).
On the other hand, in the current pandemic, efficient diagnostic methods are especially needed and help control the prevalence of the virus. Covid-19 patients are currently diagnosed using molecular and serological methods. Unlike molecular methods, serological methods have low accuracy in the early detection of patients with Covid-19 (Dhamad and Abdal Rhida, 2020). RT-qPCR is standard gold technique among all diagnostic methods. In diagnosing patients with SARS-CoV-2 infection, the combination of pharyngeal RT-qPCR and chest CT is more sensitive than other methods (Ren et al., 2020; Wei et al., 2020; Zhang et al., 2021).Several studies on the plasma of Covid-19 patients have also found that the amount of SARS-2 plasma RNA in these patients is directly related to the severity of the disease. These studies emphasize that examining the presence of SARS CoV-2 RNA and its viral load in plasma samples of patients with Covid-19 helps to predict prognosis of the disease (Veyer et al., 2020).Therefore, Covid-19 patients’ plasma analysis in terms of the presence of SARS Cov-2 RNA can be effective in better management of the covid-19 pandemic (Thijssen et al., 2020; Colagrossi et al., 2021). Furthermore, regarding the high presence of the SARS-CoV-2 genome in feces, to improve the diagnosis of the carrier, it is better to use fecal RT-qPCR as a case for hospital discharge (Ren et al., 2020; Wang et al., 2020). Several studies have confirmed the presence of the SARS-CoV-2 genome in the fecal samples of patients with Covid-19, suggesting that oral-fecal transmission of the virus can occur (Chen et al., 2020a).
Moreover, this research aimed to investigate the presence of the SARS-CoV-2 genome in the stool and plasma of Iranian patients with Covid-19, and its correlation to clinical symptoms.
2 Methods
2.1 Study Design and Patients
The enrolled patients were admitted to Taleghani Educational Hospital and Imam Hossein Educational Hospital, Shahid Beheshti University of Medical Sciences from April 6th to 15 November 2020. The study protocols were approved by the ethics committee of the Research Institute for Gastroenterology and Liver Disease (IR.SBMU.RIGLD.REC.1399.007, Tehran, Iran), and informed consent was collected from all participants.
All patients in this study were confirmed to have Covid-19 due to a positive nasopharyngeal RT-qPCR test and symptoms of pneumonia on a chest Computed tomography (CT) scan. Patients who did not have Covid-19 laboratory confirmed were not included in the study. It is noteworthy that the RT-PCR cycling threshold (Ct) for determining whether people are positive or negative for Covid-19 is a CT ≤ 40. Therefore, individuals with a CT > 40 were excluded from the study.
2.2 Sample Collection
All patients were admitted to the hospital on average 8–10 days from onset of symptoms. Fecal samples were collected from 74 registered COVID-19 patients at the beginning of admission and on the second and fourth days of the clinical course up to 3 times serially. Plasma samples were also collected from 117 other registered COVID-19 patients with the same conditions. Then all samples were stored in a special container in the freezer at −20°C. The volume of samples collected in each turn for fecal samples is between 10 and 20 ml and for plasma samples is between 5 and 10 ml.
2.3 Data Collection
191 patients demographic information, clinical characteristics (including symptoms, medical history, underlying diseases), and epidemiological, laboratory results of all patients were collected from Taleghani Educational Hospital and Imam Hossein Educational Hospital, SBMU using medical information registration system and evaluated individually.
2.4 Laboratory Assays
Plasma and feces samples were obtained and analyzed for SARS-CoV-2 RNA using RT-qPCR during the hospitalization period. Laboratory confirmation of SARS‐CoV‐2 was performed by Gastroenterology and Liver Diseases Research Center of Shahid Beheshti University of Medical Sciences in Taleghani Hospital. The nucleic acid extraction from the stool and plasma samples was performed using QIAamp Viral RNA Kit following the manufacturer’s instructions (QIAGEN, Germany). Extracted nucleic acid specimens were evaluated for SARS-CoV-2 with RT-qPCR using a SARS-CoV-2 ORF1ab/N Gene Nucleic acid detection kit (Sansure, China) and the Rotor-Gene Q real-time PCR system (QIAGEN, Germany) under manufacturer’s instructions; 4 μl 2019-nCoV-PCR-Enzyme Mix were added into 26 μl of the 2019-nCoV-PCR Mix. Then add 30 µl PCR-Master mix into PCR reaction tube with 20 µl processed sample. Reactions were incubated at 50°C for 30 min and 95°C for 1 min followed by 45 cycles at 95°C for 15 s and 60°C for the 30 s.
The severity of COVID-19 was assessed by WHO interim recommendations (World Health Organization, 2020).
2.5 Statistical Analysis
GraphPad Prism v.8.4.1 (GraphPad Software, San Diego, CA, United States) was used for all statistical analyses. The data distribution was statistically normal, according to Shapiro-Wilks test. As well as, homogeneity of variance was significant (p < 0.05), underlying that the presumption of the normality or homogeneity was met for given specimens. Therefore, student’s t-test and χ2 analysis were used to test for statistical differences among patients with negative and positive RT-qPCR tests, with a two-sided p-value of 0.05 or lower signifying significance.
3 Results
3.1 Demographics and Clinical Characteristics
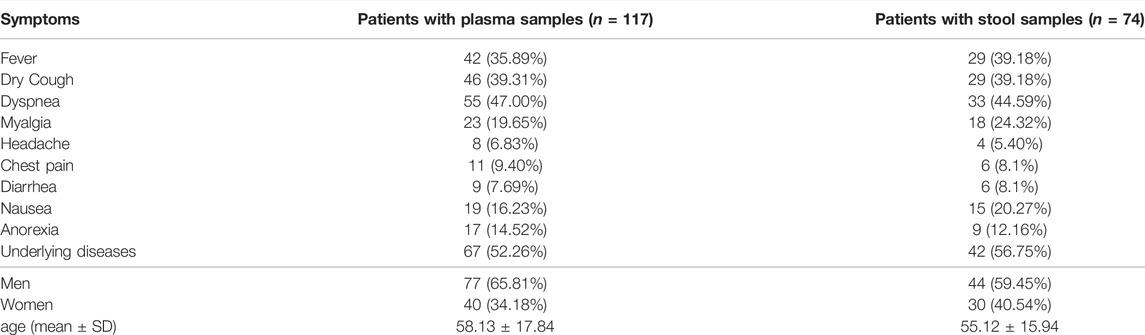
From April 6th to 15 November 2020, from the beginning of the Covid-19 pandemic to the end of the third wave of this epidemic, a total of 191 COVID‐19 hospitalized patients at Taleghani and Imam Hossein Educational Hospital, SBMU, were enrolled. Patients’ demographic and basic characteristics that were studied are listed in Table 1.
3.2 Presence of SARS-COV-2 Genome in Plasma Samples
Plasma samples of patients were collected to identify SARS-CoV-2 RNA multiple times during the hospitalization stage. As shown in Table 2, 24 of 117 (20.51%) patients analyzed positive for SARS‐CoV‐2 RNA in plasma. 12 patients were positive in the first sample (2 weeks after the onset of symptoms), four were positive in the second sample (day 3 of admission) and five patients were positive at the third sample (day 5 of admission), and other 92 (78.63%) patients tested negative. Moreover, the vital signs of patients such as heart rate, respiratory rate, temperature, and blood pressure, were recorded. 61.44 and 40.96% of patients were positive for the presence of IgG and IgM in plasma, respectively.

TABLE 2. A comparison of demographic and clinical data of patients with SARS-CoV-2 RNA positive/negative results in plasma samples.
The median (IQR) age was 58.13 ± 17.84 (18–90) years, and 40 (34.18%) of these were women. 67 (52.26%) patients had underlying diseases, including diabetes, kidney disease, cardiovascular disease, hypertension, chronic pulmonary disease, chronic kidney disease, liver disease, and cancer. The most common clinical symptoms among patients were shortness of breath (55, 47.00%), dry cough (46, 39.31%), fever (42, 35.89%), Myalgia (23.19.65%), and nausea (19, 21.56%). Less common symptoms were headache (8.6.83%), Diarrhea (9.7.69%), Chest pain (11.9.40%). Remarkably, 12 (50.00%) patients with fever and dyspnea and 16 (66.66%) patients with underlying disease tested positive for plasma RT-PCR. Also, 20 (83.33%) patients who tested positive for plasma RT-qPCR were men.
3.3 Presence of SARS-COV-2 Genome in Stool Samples
Stool samples from 74 individuals were collected and examined for the presence of SARS-COV-2 genomic RNA. Eighteen patients tested positive on the first day of admission, two patients tested positive on the third day of admission, and one patient tested positive on the fourth day of hospitalization (day 5 of admission). Table 3 shows the findings of this investigation, as well as clinical and demographic information about these individuals.
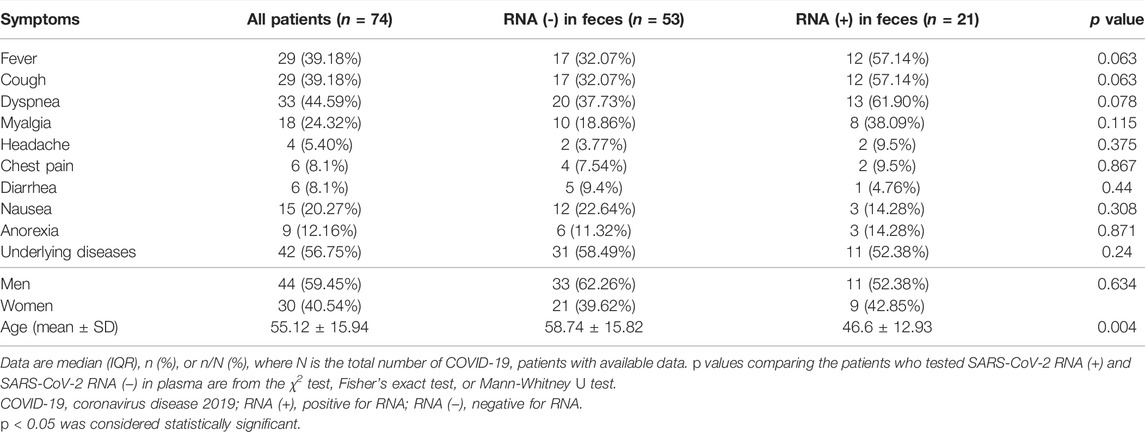
TABLE 3. A comparison of demographic and clinical data of patients with SARS-CoV-2 RNA positive/negative results in stool samples.
44 (59.45%) of these patients were male, 30 (40.54%) were female and the mean age of these was 55.12 ± 15.94 (24–86) years. 42 (56.75%) patients had an underlying disease, including diabetes, kidney disease, cardiovascular disease, hypertension, chronic pulmonary disease, chronic kidney disease, liver disease, and cancer.
Less common symptoms were headache (4.5.40%), chest pain, and diarrhea (6.8.1%). The most common clinical symptoms among patients were fever (6.42.85%), anorexia (9.12.16%), nausea (15.20.27%). Stool samples were collected to detect COVID-19 RNA several times during the hospitalization stage. As shown in Table 3, 21 of 74 (28.37%) patients tested positive for SARS‐CoV‐2 RNA in feces, and the other 53 (71.62%) patients tested negative. It is important to note that all those who had nausea tested positive for SARS‐CoV‐2 RNA RT-PCR (p value = 0.308).
3.4 Laboratory and Vital Finding
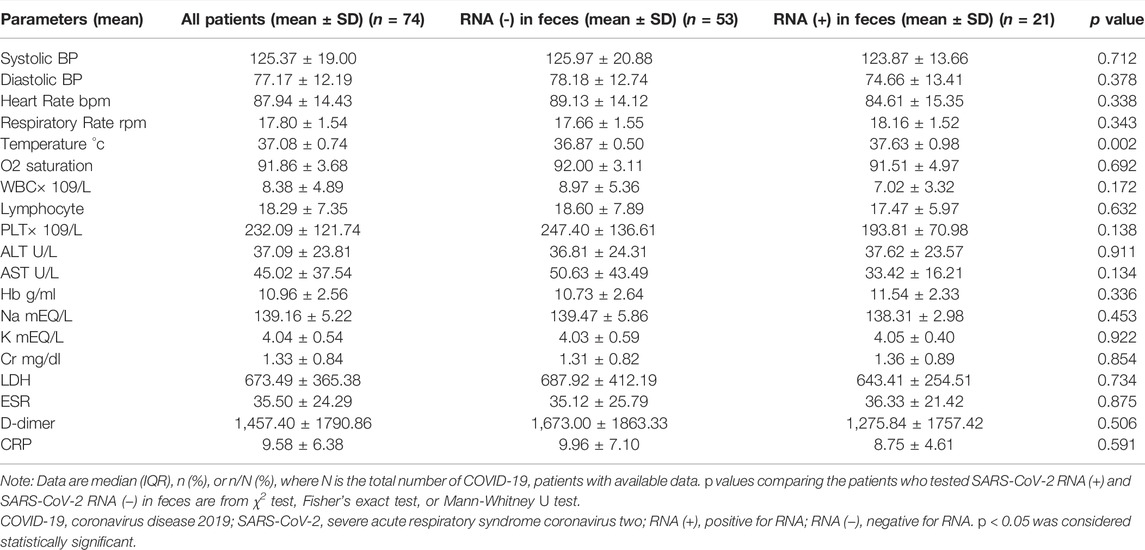
The laboratory and vital data of all patients whose plasma samples were evaluated for the presence of SARS COV-2 RNA are recorded in Table 4. Compared to people whose plasma SARS‐CoV‐2 RNA RT-qPCR test results are negative, the average amount of Na (p value = 0.333) and d-dimer (p value = 0.270) in the blood of people whose plasma SARS‐CoV‐2 RNA RT-qPCR test results are positive are higher, and this difference is not significant.
Besides, laboratory findings and vital data of patients whose stools sample were evaluated for the presence of SARS‐CoV‐2 RNA by RT-qPCR test, are shown in Table 5, which showed the mean temperature of patients whose stool samples were positive for SARS-CoV-2 RNA was higher than that of patients whose stool samples were negative for SARS-CoV-2 RNA, and this difference was significant.
3.5 Correlation Between Disease Severity and Viral Load in Stool and Plasma Samples
Figure 2 indicates the severity of COVID-19. In general, 44 (37.60%) of patients whose plasma samples were examined, belonged to the severe clinical group. 20% of Severe patients were positive for the presence of RNA virus in plasma samples. Furthermore, 27 (36.48%) of the patients whose stool samples were analyzed and classified as severe, with 46.66% testing positive for an RNA virus.
In general, 28.37% of fecal samples and 20.51% of plasma samples were positive for SARS‐CoV‐2 RNA and their viral load was evaluated. Viral load was significantly different (p < 0.05) in plasma samples compared to stool samples. Plasma samples showed higher viral load than stool samples (Figure 1). Viral loads in plasma and fecal samples did not show a significant difference among the patients with mild disease and patients with severe disease (Figure 2).
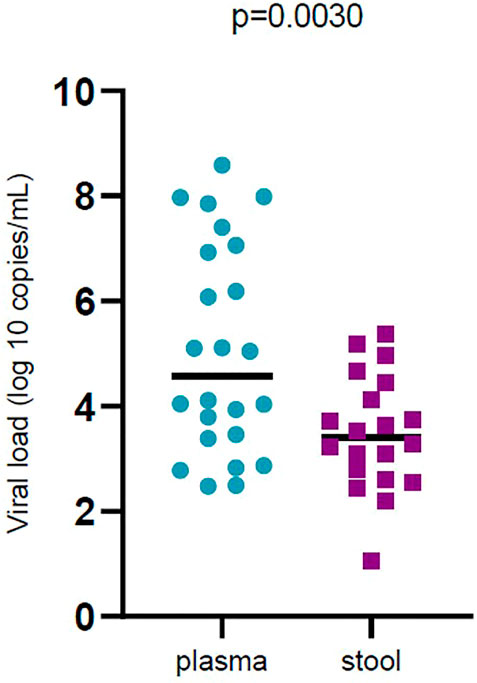
FIGURE 1. Comparison of severe acute respiratory syndrome coronavirus 2 (SARS -CoV-2) viral load by sample types (plasma and stool). Black bars represent medians.
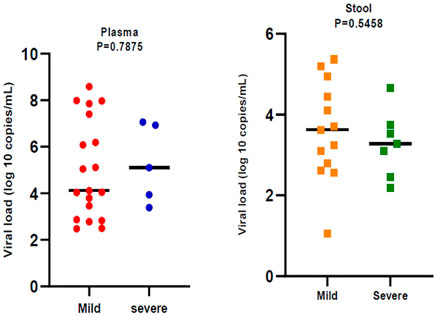
FIGURE 2. Comparison of severe acute respiratory syndrome coronavirus 2 (SARS -CoV-2) viral load by disease severity. Black bars represent medians.
4 Discussion
The lungs are primary SARS-CoV-2’s target organ. The respiratory system is also recognized to be the virus’s most prevalent route of transmission. SARS-CoV-2 penetrates target cells by attaching to the ACE-2 receptor, and since this receptor is found on the surface of other organ cells as well as lung cells, there’s a chance it might infect cells other than lung cells. Therefore, transmission routes other than the respiratory route should also be considered (Robba et al., 2020; Trypsteen et al., 2020; Vinayagam and Sattu, 2020). However, using multiple samples at the same time could prevent probable viral transmission through other routes like the oral-fecal and other body fluids (Wang et al., 2020). Moreover, RNA shedding in different biological samples and recognizing its transmission routes in terms of biosafety is essential for health care workers and patient management in hospitals (Eastin and Eastin, 2020). Furthermore, for a more accurate diagnosis of Covid-19 patients and reduce viral shedding, it is necessary to analyze the association between the severity of illness and the presence of the virus in different samples of patients.
The presence of pathogens in various samples of other viruses was considered. For example, Hepatitis C virus RNA, has been found in the saliva, bile, and feces of chronic hepatitis C patients. Patients with a higher serum HCV viral load had more positive saliva samples. HCV RNA was reported to be higher in the feces of patients with low platelets, in addition to the presence of the virus genome in the feces of men more than women. (Yanaga et al., 1997; Suzuki et al., 2005; Wang et al., 2006; Heidrich et al., 2016). Furthermore, in studies on the Ebola virus, genomic RNA was found in a variety of bodily fluids, including urine, feces, blood, sweat, saliva, tears, cerebrospinal fluid, amniotic fluid, breast milk, and sperm, in acute Ebola virus disease patients (Rodriguez et al., 1999; Bausch et al., 2007; Feldmann and Geisbert, 2011; Sagui et al., 2015; INTERIM GUIDANCE, 2016). In a study, it was stated that the viral load in the feces of Ebola patients peaked during the disease (Prescott et al., 2015; Schibler et al., 2015). Also in a study conducted on plasma, urine, and saliva samples of patients with dengue virus by Andris et al., 85.4% of plasma samples, 41.6% of urine samples, and 39% of saliva samples in terms of The presence of dengue virus genome were positive (Andries et al., 2015). According to a study by Chan et al., SARS-CoV RNA is detected only in feces after day 5, and the proportion of positive stool samples increases until day eleven when it reaches its peak (Chan et al., 2004).
The presence of the viral genome in different clinical samples is highly important and its relationship with the severity of clinical symptoms, this study was designated to investigate the presence of SARS-CoV-2 genome in stool and plasma samples of patients with Covid-19 and to study their association with clinical signs of individuals. Thus, our result showed stool samples from 74 patients and plasma samples from 117 patients with Covid-19 were evaluated. 27.03% of stool samples and 20.51% of plasma specimens were positive for the SARS-CoV-2 genome.
Many diagnostic procedures have been explored to detect infected patients since the commencement of the Covid-19 outbreak. Molecular assays for the identification of the SARS-CoV-2 genome or serological testing for the presence of antibodies to the virus are used to identify Covid-19 patients (Jarrom et al., 2020). Methods such as RT-qPCR, which identifies the viral nucleic acid, are frequently used to detect the presence of the virus. For the clinical diagnosis of SARS-CoV-2, the RT-qPCR of the nasopharyngeal swab and chest CT scan is commonly used. The sensitivity of RT-qPCR tests to identify Covid-19 patients was found to be 88% in research by Burnheim et al. (Bernheim et al., 2020). The RT-qPCR test was negative in a percentage of individuals whose Covid-19 was approved, according to Lee et al. (Li et al., 2020a). This might be due to a sample deficit, a laboratory mistake, or a lack of virus particles in the sample Regarding the prevalence of COVID-19 and the high potential of SARS-CoV-2 transmission via non-respiratory routes, also due to the high viral load in these samples, studying different samples for viral genome to reduce virus prevalence is significant.
Considering the importance of the issues raised, in research on samples from laboratory-confirmed COVID-19 patients, Chen et al. found that 88.2 percent of pharyngeal specimens, 11.5% of plasma specimens, and 21.2% of fecal specimens were positive for SARS-CoV-2 RNA (Chan et al., 2020). In another research, 59% of stool samples, 41% of plasma samples, and 1% of urine samples were positive for the COVID-19. The viral load of respiratory samples was stronger than in other samples, according to Zheng et al. (Jarrom et al., 2020). Lamers et al. Have presented evidence of contamination and proliferation of SARS-Cov-2 in small intestinal enterocytes, which increases the likelihood of infection and proliferation of these particles in human intestinal enterocytes (Lamers et al., 2020).Wu et al. (Bernheim et al., 2020) showed that the SARS-COV-2 RNA was present in the laboratory-confirmed COVID-19 patients’ fecal samples to 5 weeks after the negative nasopharyngeal test. Although no confirmation of SARS-CoV-2 particles being transmitted through feces has been found, several studies have raised concerns about the shedding of viral active particles through patients’ feces (Lamers et al., 2020; Mirjalali et al., 2020; Tian et al., 2020).
Chou et al. stated in a research on the transmission of SARS-CoV-2 via blood products that the likelihood of transmission through this route should be examined (Cho et al., 2020). Meanwhile, the European Center for Disease Control and Prevention (ECDC) suggests delaying blood donations for 21 days after any probable contact with authorized patients in order to prevent the virus from spreading via blood products (Risk Assessment, 2020). As a result, the role of blood products in the transmission of the SARS-COV-2 is important.
Although it is critical to assess non-respiratory samples such as stools and blood to prevent the virus from spreading in pandemics and to ensure the safety of healthcare personnel, it is critical to examine the relationship among clinical signs or laboratory data and the presence of the SARS-CoV-2 genome in various samples.
For example, Chen et al. Found that The severity of COVID-19 pneumonia was positively correlated with plasma CRP levels that a non-specific reactive protein that increased in infection and inflammation. High levels of this protein have been observed in the plasma of Covid-19 patients, which can be used for diagnosis and prognosis (Chen et al., 2020b). In our study, people who had a plasma positive for SARS-CoV-2 RNA had a higher CRP than people who did not have SARS-CoV-2 RNA. This can increase the possibility of the presence of an active virus and can also be used to diagnose and prognosis.
As well, because fever in patients is a sign of active immunity (Moltz, 1993; Young and Saxena, 2014), and according to this study’s findings, the temperature in patients whose SARS-CoV-2 RNA was detected in their feces is higher than patients whose feces are negative for SARS-CoV-2 RNA, the possibility of SARS-CoV-2 active particles exists in these patients is increased.
There is debate as to whether SARS-CoV-2 shedding in stool and plasma samples is associated with the severity of the disease. Some studies found no link, which is consistent with the findings of our research (Chen et al., 2020a). Other research has shown a link between SARS-CoV-2 shedding in stool and plasma samples and the severity of COVID-19 symptoms in humans (Bermejo-Martin et al., 2020; Li et al., 2020b; Reuken et al., 2020; Shang et al., 2020).
No association was found between disease severity and gastrointestinal symptoms with the presence of SARS-CoV-2 RNA and the possible presence of the virus in the fecal and plasma samples of Covid-19 patients. This difference among studies may be in terms of environmental and genetic differences affecting clinical signs in the course of the disease, and differences in terms of SARS-CoV-2 strains.
Our study was performed on limited patients. more specimens better define viral shedding in plasma and feces and their association with clinical symptoms, and the usefulness of recommending routine testing of non-respiratory specimens.
The deterministic correlation between plasma and fecal viral shedding and the severity of gastrointestinal symptoms has not yet been determined, but it appears that SARS-CoV-2 may be present in the gut without affecting the severity of gastrointestinal symptoms.
Although SARS-CoV-2 RNA was detected in the feces of Covid-19 patients, further studies are needed to prove or disprove its infectious potential.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the IR.SBMU.RIGLD.REC.1399.007, Tehran, Iran. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SM, SH, and MZ conceived the study, HA, AY, SB, SSHA, MMO, and HM performed the sample collection, MMA, MN, SG, and SM carried out the laboratory and molecular tests, MMA, SSHO, and SM carried out the interpretation and analyze of the data, MMA, SSHO, SM, and SH drafted the manuscript, HA, SSHA, and MZ critically revised the manuscript for intellectual content. All authors read and approved the final manuscript.
Funding
This research was supported by the Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran (IR-RIGLD-1110.2).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to the Institute laboratory staff for their valuable assistance, especially Shabnam Kazemian.
References
Andries, A.-C., Duong, V., Ly, S., Cappelle, J., Kim, K. S., Lorn Try, P., et al. (2015). Value of Routine Dengue Diagnostic Tests in Urine and Saliva Specimens. PLoS Negl. Trop. Dis. 9 (9), e0004100. doi:10.1371/journal.pntd.0004100
Bausch, D. G., Towner, J. S., Dowell, S. F., Kaducu, F., Lukwiya, M., Sanchez, A., et al. (2007). Assessment of the Risk of Ebola Virus Transmission from Bodily Fluids and Fomites. J. Infect. Dis. 196 (Suppl. ment_2), S142–S147. doi:10.1086/520545
Bermejo-Martin, J. F., González-Rivera, M., Almansa, R., Micheloud, D., Tedim, A. P., Domínguez-Gil, M., et al. (2020). Viral RNA Load in Plasma Is Associated with Critical Illness and a Dysregulated Host Response in COVID-19. Crit. Care 24 (1), 691–713. doi:10.1186/s13054-020-03398-0
Bernheim, A., Mei, X., Huang, M., Yang, Y., Fayad, Z. A., Zhang, N., et al. (2020). Chest CT Findings in Coronavirus Disease-19 (COVID-19): Relationship to Duration of Infection. Radiology 295, 200463. doi:10.1148/radiol.2020200463
Chan, J. F., Yip, C. C., To, K. K., Tang, T. H., Wong, S. C., Leung, K. H., et al. (2020). Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J. Clin. Microbiol. 58 (5), e00310–20. doi:10.1128/JCM.00310-20
Chan, K. H., Poon, L. L. L. M., Cheng, V. C. C., Guan, Y., Hung, I. F. N., Kong, J., et al. (2004). Detection of SARS Coronavirus in Patients with Suspected SARS. Emerg. Infect. Dis. 10 (2), 294–299. doi:10.3201/eid1002.030610
Chen, W., Zheng, K. I., Liu, S., Yan, Z., Xu, C., and Qiao, Z. (2020). Plasma CRP Level Is Positively Associated with the Severity of COVID-19. Ann. Clin. Microbiol. Antimicrob. 19 (1), 18–27. doi:10.1186/s12941-020-00362-2
Chen, Y., Chen, L., Deng, Q., Zhang, G., Wu, K., Ni, L., et al. (2020). The Presence of SARS‐CoV‐2 RNA in the Feces of COVID‐19 Patients. J. Med. Virol. 92 (7), 833–840. doi:10.1002/jmv.25825
Cheung, K. S., Hung, I. F. N., Chan, P. P. Y., Lung, K. C., Tso, E., Liu, R., et al. (2020). Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples from a Hong Kong Cohort: Systematic Review and Meta-Analysis. Gastroenterology 159 (1), 81–95. doi:10.1053/j.gastro.2020.03.065
Cho, H. J., Koo, J. W., Roh, S. K., Kim, Y. K., Suh, J. S., Moon, J. H., et al. (2020). COVID-19 Transmission and Blood Transfusion: A Case Report. J. Infect. Public Health 13 (11), 1678–1679. doi:10.1016/j.jiph.2020.05.001
Colagrossi, L., Antonello, M., Renica, S., Merli, M., Matarazzo, E., Travi, G., et al. (2021). SARS-CoV-2 RNA in Plasma Samples of COVID-19 Affected Individuals: a Cross-Sectional Proof-Of-Concept Study. BMC Infect. Dis. 21 (1), 184–186. doi:10.1186/s12879-021-05886-2
Dhamad, A. E., and Abdal Rhida, M. A. (2020). COVID-19: Molecular and Serological Detection Methods. PeerJ 8, e10180. doi:10.7717/peerj.10180
Eastin, C., and Eastin, T. (2020). Epidemiological Characteristics of 2143 Pediatric Patients with 2019 Coronavirus Disease in China. J. Emerg. Med. 58 (4), 712–713. doi:10.1016/j.jemermed.2020.04.006
Feldmann, H., and Geisbert, T. W. (2011). Ebola Haemorrhagic Fever. Lancet 377 (9768), 849–862. doi:10.1016/s0140-6736(10)60667-8
Heidrich, B., Steinmann, E., Plumeier, I., Kirschner, J., Sollik, L., Ziegert, S., et al. (2016). Frequent Detection of HCV RNA and HCVcoreAg in Stool of Patients with Chronic Hepatitis C. J. Clin. Virology 80, 1–7. doi:10.1016/j.jcv.2016.04.006
INTERIM GUIDANCE (2016). Clinical Care for Survivors of Ebola Virus Disease. Geneva: World Health Organization.
Jarrom, D., Elston, L., Washington, J., Prettyjohns, M., Cann, K., Myles, S., et al. (2020). Effectiveness of Tests to Detect the Presence of SARS-CoV-2 Virus, and Antibodies to SARS-CoV-2, to Inform COVID-19 Diagnosis: a Rapid Systematic Review. BMJ evidence-based Med. 27, 33–45. doi:10.1136/bmjebm-2020-111511
Lamers, M. M., Beumer, J., Van Der Vaart, J., Knoops, K., Puschhof, J., Breugem, T. I., et al. (2020). SARS-CoV-2 Productively Infects Human Gut Enterocytes. Science 369 (6499), 50–54. doi:10.1126/science.abc1669
Li, X.-Y., Dai, W.-J., Wu, S.-N., Yang, X.-Z., and Wang, H.-G. (2020). The Occurrence of Diarrhea in COVID-19 Patients. Clin. Res. hepatology gastroenterology 44 (3), 284–285. doi:10.1016/j.clinre.2020.03.017
Li, Y., Yao, L., Li, J., Chen, L., Song, Y., Cai, Z., et al. (2020). Stability Issues of RT‐PCR Testing of SARS‐CoV‐2 for Hospitalized Patients Clinically Diagnosed with COVID‐19. J. Med. Virol. 92 (7), 903–908. doi:10.1002/jmv.25786
Lippi, G., Sanchis-Gomar, F., and Henry, B. M. (2020). Coronavirus Disease 2019 (COVID-19): the Portrait of a Perfect Storm. Ann. Transl. Med. 8 (7), 497. doi:10.21037/atm.2020.03.157
Mirjalali, H., Nazemalhosseini-Mojarad, E., Yadegar, A., Mohebbi, S. R., Baghaei, K., Shahrokh, S., et al. (2020). The Necessity of Stool Examination in Asymptomatic Carriers as a Strategic Measure to Control Further Spread of SARS-CoV-2. Front. Public Health 8, 553589. doi:10.3389/fpubh.2020.553589
Moltz, H. (1993). Fever: Causes and Consequences. Neurosci. Biobehav. Rev. 17 (3), 237–269. doi:10.1016/s0149-7634(05)80009-0
Prescott, J., Bushmaker, T., Fischer, R., Miazgowicz, K., Judson, S., and Munster, V. J. (2015). Postmortem Stability of Ebola Virus. Emerg. Infect. Dis. 21 (5), 856–859. doi:10.3201/eid2105.150041
Ren, X., Liu, Y., Chen, H., Liu, W., Guo, Z., Chen, C., et al. (2020). “Application and Optimization of RT-PCR in Diagnosis of SARS-CoV-2 Infection,” in Chaoqun and Zhou, Jianhui and Xiao, Qiang and Jiang, Guan-Min and Shan, Hong, Application and Optimization of RT-PCR in Diagnosis of SARS-CoV-2 Infection (2/25/2020).
Reuken, P. A., Wüst, M., Löffler, B., Bauer, M., and Stallmach, A. (2020). Letter: SARS-CoV-2-Induced Gastrointestinal Inflammation. Aliment. Pharmacol. Ther. 52 (11-12), 1748–1749. doi:10.1111/apt.16087
Risk Assessment (2020). Outbreak of Acute Respiratory Syndrome Associated with a Novel Coronavirus, Wuhan, China; First Update 22 January 2020. Stockholm: ECDC.
Robba, C., Battaglini, D., Pelosi, P., and Rocco, P. R. M. (2020). Multiple Organ Dysfunction in SARS-CoV-2: MODS-CoV-2. Expert Rev. Respir. Med. 14 (9), 865–868. doi:10.1080/17476348.2020.1778470
Rodriguez, L. L., De Roo, A., Guimard, Y., Trappier, S. G., Sanchez, A., Bressler, D., et al. (1999). Persistence and Genetic Stability of Ebola Virus during the Outbreak in Kikwit, Democratic Republic of the Congo, 1995. J. Infect. Dis. 179 (Suppl. ment_1), S170–S176. doi:10.1086/514291
Sagui, E., Janvier, F., Baize, S., Foissaud, V., Koulibaly, F., Savini, H., et al. (2015). Severe Ebola Virus Infection with Encephalopathy: Evidence for Direct Virus Involvement. Clin. Infect. Dis. 61 (10), 1627–1628. doi:10.1093/cid/civ606
Schibler, M., Vetter, P., Cherpillod, P., Petty, T. J., Cordey, S., Vieille, G., et al. (2015). Clinical Features and Viral Kinetics in a Rapidly Cured Patient with Ebola Virus Disease: a Case Report. Lancet Infect. Dis. 15 (9), 1034–1040. doi:10.1016/s1473-3099(15)00229-7
Shang, H., Bai, T., Chen, Y., Huang, C., Zhang, S., Yang, P., et al. (2020). Outcomes and Implications of Diarrhea in Patients with SARS-CoV-2 Infection. Scand. J. gastroenterology 55 (9), 1049–1056. doi:10.1080/00365521.2020.1800078
Suzuki, T., Omata, K., Satoh, T., Miyasaka, T., Arai, C., Maeda, M., et al. (2005). Quantitative Detection of Hepatitis C Virus (HCV) RNA in Saliva and Gingival Crevicular Fluid of HCV-Infected Patients. J. Clin. Microbiol. 43 (9), 4413–4417. doi:10.1128/jcm.43.9.4413-4417.2005
Thijssen, M., Devos, T., Ejtahed, H.-S., Amini-Bavil-Olyaee, S., Pourfathollah, A. A., and Pourkarim, M. R. (2020). Convalescent Plasma against COVID-19: a Broad-Spectrum Therapeutic Approach for Emerging Infectious Diseases. Microorganisms 8 (11), 1733. doi:10.3390/microorganisms8111733
Tian, Y., Rong, L., Nian, W., and He, Y. (2020). Review Article: Gastrointestinal Features in COVID-19 and the Possibility of Faecal Transmission. Aliment. Pharmacol. Ther. 51 (9), 843–851. doi:10.1111/apt.15731
Trypsteen, W., Van Cleemput, J., Snippenberg, W. v., Gerlo, S., and Vandekerckhove, L. (2020). On the Whereabouts of SARS-CoV-2 in the Human Body: A Systematic Review. PLoS Pathog. 16 (10), e1009037. doi:10.1371/journal.ppat.1009037
Udugama, B., Kadhiresan, P., Kozlowski, H. N., Malekjahani, A., Osborne, M., Li, V. Y. C., et al. (2020). Diagnosing COVID-19: the Disease and Tools for Detection. ACS Nano 14 (4), 3822–3835. doi:10.1021/acsnano.0c02624
Veyer, D., Kernéis, S., Poulet, G., Wack, M., Robillard, N., Taly, V., et al. (2020). Highly Sensitive Quantification of Plasma Severe Acute Respiratory Syndrome Coronavirus 2 RNA Sheds Light on its Potential Clinical Value. Clin. Infect. Dis. 73 (9), e2890–e2897. doi:10.1093/cid/ciaa1196
Vinayagam, S., and Sattu, K. (2020). SARS-CoV-2 and Coagulation Disorders in Different Organs. Life Sci. 260, 118431. doi:10.1016/j.lfs.2020.118431
Wang, C. C., Morishima, C., Chung, M., Engelberg, R., Krantz, E., Krows, M., et al. (2006). High Serum Hepatitis C Virus (HCV) RNA Load Predicts the Presence of HCV RNA in Saliva from Individuals with Chronic and Acute HCV Infection. J. Infect. Dis. 193 (5), 672–676. doi:10.1086/499602
Wang, W., Xu, Y., Gao, R., Lu, R., Han, K., Wu, G., et al. (2020). Detection of SARS-CoV-2 in Different Types of Clinical Specimens. Jama 323 (18), 1843–1844. doi:10.1001/jama.2020.3786
Wei, J., Xu, H., Xiong, J., Shen, Q., Fan, B., Ye, C., et al. (2020). 2019 Novel Coronavirus (COVID-19) Pneumonia: Serial Computed Tomography Findings. Korean J. Radiol. 21 (4), 501–504. doi:10.3348/kjr.2020.0112
World Health Organization (2020). Laboratory Testing for Coronavirus Disease ( COVID-19) in Suspected Human Cases: Interim Guidance, 19 March 2020. World Health Organization.
Wu, A., Peng, Y., Huang, B., Ding, X., Wang, X., Niu, P., et al. (2020). Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell host microbe 27 (3), 325–328. doi:10.1016/j.chom.2020.02.001
Yanaga, K., Yoshizumi, T., Uchiyama, H., Okano, S., Takenaka, K., and Sugimachi, K. (1997). Detection of Hepatitis C Virus RNA in Bile. Am. J. Gastroenterol. 92 (10), 1927–1928. doi:10.1007/978-3-319-03746-2_1
Young, P., and Saxena, M. (2014). “Fever Management in Intensive Care Patients with Infections,” in Annual Update in Intensive Care and Emergency Medicine 2014, 3–16. doi:10.1007/978-3-319-03746-2_1
Zhang, J.-F., Liu, J., Ma, H.-N., Feng, K., Chen, Z.-W., Yang, L.-S., et al. (2021). RT-PCR Combined with CT Examination in the Diagnosis and Prognosis Evaluation of COVID-19 Patients in Fangcang Hospital: a Case Series. J. Multidiscip. Healthc. 14, 145–149. doi:10.2147/jmdh.s293601
Keywords: SARS-CoV-2, COVID-19, RT-qPCR, plasma, feces, COVID-19 nucleic acid testing
Citation: Makhmalbaf M, Hosseini SM, Aghdaei HA, Niasar MS, Shoraka S, Yadegar A, Baradaran Ghavami S, Shahrokh S, Moshari M, Malekpour H, Zali MR and Mohebbi SR (2022) Detection of SARS-CoV-2 Genome in Stool and Plasma Samples of Laboratory Confirmed Iranian COVID-19 Patients. Front. Mol. Biosci. 9:865129. doi: 10.3389/fmolb.2022.865129
Received: 29 January 2022; Accepted: 10 June 2022;
Published: 28 June 2022.
Edited by:
Charareh Pourzand, University of Bath, United KingdomReviewed by:
Mohammad Mozafarihashjin, University of Toronto, CanadaMahmoudReza Pourkarim, KU Leuven, Belgium
Copyright © 2022 Makhmalbaf, Hosseini, Aghdaei, Niasar, Shoraka, Yadegar, Baradaran Ghavami, Shahrokh, Moshari, Malekpour, Zali and Mohebbi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seyed Reza Mohebbi, sr.mohebbi@sbmu.ac.ir
 Mobin Makhmalbaf
Mobin Makhmalbaf Seyed Masoud Hosseini2
Seyed Masoud Hosseini2  Hamid Asadzadeh Aghdaei
Hamid Asadzadeh Aghdaei Shahrzad Shoraka
Shahrzad Shoraka Abbas Yadegar
Abbas Yadegar Shaghayegh Baradaran Ghavami
Shaghayegh Baradaran Ghavami Shabnam Shahrokh
Shabnam Shahrokh Seyed Reza Mohebbi
Seyed Reza Mohebbi