- 1Wuhan Institute of Biomedical Sciences, School of Medicine, Jianghan University, Wuhan, China
- 2Hepatic Disease Institute, Hubei Key Laboratory of Theoretical and Applied Research of Liver and Kidney in Traditional Chinese Medicine, Hubei Provincial Hospital of Traditional Chinese Medicine, Wuhan, China
- 3Hepatic Disease Institute, Hubei Province Academy of Traditional Chinese Medicine, Wuhan, China
SARS-CoV-2 is the causative agent for the global COVID-19 pandemic; however, the interaction between virus and host is not well characterized. Natural killer cells play a key role in the early phase of the antiviral response, and their primary functions are dependent on signaling through the killer cell immunoglobulin-like receptor (KIR). This study measured the association between KIR/HLA class I ligand pairings and the occurrence and development of COVID-19. DNA of blood samples from 257 COVID-19 patients were extracted and used to detect KIR and HLA-C gene frequencies using single strain sequence-specific primer (SSP) PCR. The frequency of these genes was compared among 158 individuals with mild COVID-19, 99 with severe disease, and 98 healthy controls. The frequencies of KIR2DL2 (P=0.04, OR=1.707), KIR2DS3 (P=0.047, OR=1.679), HLA-C1C1 (P<0.001, OR=3.074) and the KIR2DL2/HLA-C1C1 pairing (P=0.038, OR=2.126) were significantly higher in the COVID-19 patients than the healthy controls. At the same time, the frequency of KIR2DL3+KIR2DL2-/HLA-C1+Others+ was lower in COVID-19 patients than in healthy individuals (P=0.004, OR=0.477). These results suggest that the protective effect of KIR2DL3 against SARS-CoV-2 infection is related to the absence of the KIR2DL2 gene. This study found no correlation between the frequencies of these genes and COVID-19 pathogenesis. Global statistical analysis revealed that the incidence of COVID-19 infection was higher in geographic regions with a high frequency of KIR2DL2. Together these results suggest that the KIR2DL2/HLA-C1C1 gene pairing may be a risk factor for SARS-CoV-2 infection.
Introduction
On January 30, 2020, the World Health Organization (WHO) declared the 2019 Coronavirus Disease (COVID-19) outbreak an important public health event (1). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a coronaviruses family member, is characterized by a single-stranded RNA with positive sense. The coronavirus genome is highly susceptible to mutations, resulting in genetic drift and allowing the virus to evade immune recognition (2). The virus is still circulating worldwide, and the new coronavirus disease caused by SARS-CoV-2 is a public health crisis that requires an active response (3). Studies of SARS-CoV-2 infection have shown that this infection spreads easily among immediate family members. The speed of viral transmission and the pathogenicity of this disease differ by geographic region and while this is related to differences in the implementation of prevention and control strategies, the occurrence and progression of COVID-19 may also be related to the host’s genetic background (4).
Human leukocyte antigen (HLA) polymorphism is a genetic factor associated with variations in susceptibility to virus infection. For example, SARS-CoV-2 patients in Saudi Arabia were found to have a higher frequency of HLA-A*01, B*56, and C*01 than uninfected individuals, and the frequency of HLA-A*03 and C*06 was particularly high in patients who died of COVID-19 (5). In a Japanese study, HLA-A*11, HLA-C*12, and HLA-B*52 correlated significantly with disease severity (6). In Italy, COVID-19 infection is typically more severe in the northern region and milder in the southern region. The regional distribution of HLA-C*07 in the Italian population is positively correlated with COVID-19 morbidity and mortality (7). SARS-CoV-2 infection disrupts immune homeostasis, leading to an imbalance of the immune regulatory system. Infection can also result in a decline in macrophages and NK cells (8–10).
NK cells are important lymphocytes of the innate immune response that can directly kill target cells without damaging healthy cells (11). This cell type is relatively abundant, accounting for 5–20% of lymphocytes (12), and plays a key role in both antiviral and antitumor immunity. NK cell activation is dependent on signals triggered by an array of inhibitory and activating receptors that ensure self-tolerance against healthy cells while destroying virally infected target cells. The killer cell immunoglobulin-like receptor (KIR), an immunoglobulin superfamily member, is one of the important surface receptors involved in NK cell activation. KIRs are expressed on the surface of NK cells and some T lymphocytes (13). NK cells account for about 15% of cells in the human lung and CD56dim NK cell subsets expressing KIR and other receptors account for 60–80% of this population (14). To date, 16 distinct KIR gene loci, including the KIR2DP1 and KIR3DP1 pseudogenes, have been identified, which has created considerable diversity in the number and type of KIR expressed on the NK cell surface. Many KIR ligands have been identified, including KIR3DL1 combined with HLA-Bw4 (15), KIR2DL1 combined with HLA-Cw2, HLA-Cw4, HLA-Cw5 or HLA-Cw6 (called C2), and KIR2DL2, KIR2DL3 and KIR2DS2 combined with HLA-Cw1, HLA-Cw3, HLA-Cw7 or HLA-Cw8 (called C1) (16). Based on the polymorphisms at positions 77 and 80 in the HLA heavy chain a1 domain, KIR2DL1, KIR2DL2/3, and KIR2DS2 recognize different HLA-C allotypes. In Caucasians, patients with persistent hepatitis C virus (HCV) infection have a higher frequency of KIR2DL3 than those who have successfully cleared infection (17). KIR2DL3 is also closely related to the occurrence of fatal cerebral malaria (18) and KIR3DS1 is associated with the delayed development of AIDS and clearance of HCV (19).
The individual immune response is closely related to the KIR/HLA-C pairing. At the same time, viral replication and pathogenicity are associated with the genetic background of the host. This study assessed the frequency of the KIR and HLA-C genes in COVID-19 patients infected in the early phase of the pandemic and healthy people living in Wuhan, China, and identified specific protective or susceptible KIR/HLA-C gene pairings.
Materials and Methods
Study Subjects
This study conformed to the 1975 Declaration of Helsinki guidelines and permission was obtained from the Ethics Committee of Jianghan University and Hubei Provincial Hospital of Traditional Chinese Medicine. From January to October 2020, the research team recruited 98 healthy volunteers who were not infected with SARS-CoV-2 and 257 COVID-19 patients who received rehabilitation treatment at the Hubei Provincial Hospital of Traditional Chinese Medicine in Wuhan, China to conduct a retrospective study. All participants were Chinese Han.
COVID-19 patients were divided into a group of 158 individuals with mild infection and a group of 99 people with severe infection using the Guidelines on the Diagnosis and Treatment of Novel Coronavirus issued by the National Health Commission in China. In brief, mild disease was defined as a lack of respiratory symptoms and no pulmonary radiological manifestations, or mild respiratory symptoms and radiological evidence of pneumonia. Severe cases were defined as SpO2 ≤93% that required oxygen support or requiring heart-lung machine support for acute respiratory distress syndrome. All study participants were aware of the research content and met the ethical requirements.
DNA Extraction and Genotyping
Genomic DNA was extracted from 5 ml EDTA anticoagulated peripheral blood using an SE Blood DNA Kit (Omega Bio-Tek Inc, USA). Fifty forward and reverse primer pairs were used to specifically amplify the KIR gene by sequence-specific primer (SSP) PCR (20). SSP PCR was performed according to a method based on the HLA-C allele that was described previously (21). After separation on a 1.5% agarose gel, it was determined whether there was a PCR product. All gels were assessed by two independent observers using a DNA gel imaging system and if any inconsistencies arose, the samples were re-typed.
Statistical Methods
Using SPSS 18.0 software for Windows, KIR/HLA genotype frequencies were compared between healthy controls and COVID-19 patients using the Chi-square test, p-values, and Odds ratios (ORs). ORs and 95% confidence intervals (CIs) were calculated using the quantitative data from COVID-19 patients and healthy controls. The 95% CI confidence interval for a meaningful OR value excludes the value of 1. The test data is only considered statistically significant if both the p-value and the OR value of the gene or paired combination frequency are statistically significant. The same method was used to analyze the frequencies of related genes in COVID-19 patients with mild and severe disease.
Results
Participant Recruitment and Testing
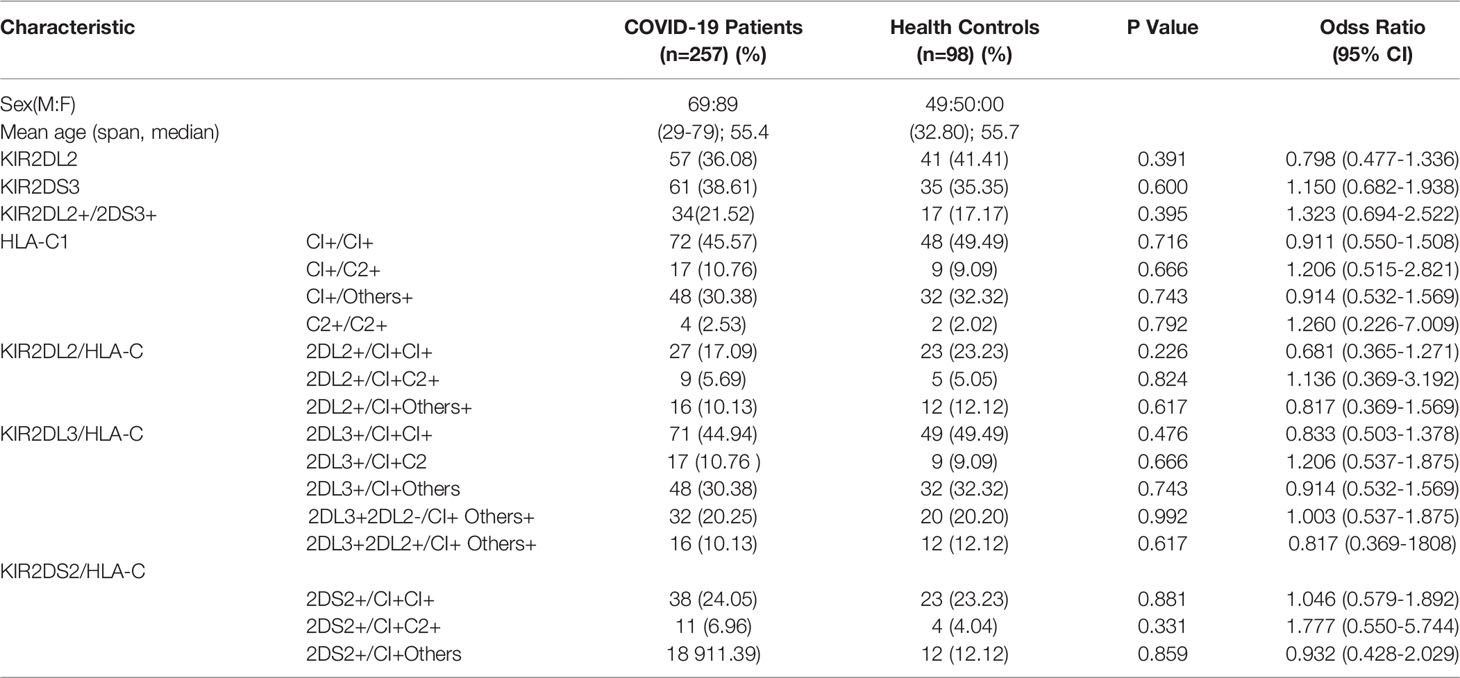
The study subjects were all from the Hubei Provincial Hospital of Traditional Chinese Medicine. Ninety-eight healthy volunteers (59 males and 39 females) were verified as being COVID-19 negative using nucleic acid testing and medical history review and had no history of HIV, HBV, or HCV infection. All volunteers lived in Wuhan during the COVID-19 outbreak. The 257 COVID-19 patients included 158 (69 men and 89 women) with mild disease and 99 (49 men and 50 women) with severe disease who were diagnosed with COVID-19 using a nucleic acid test, CT scan, and other routine tests. After recovery, the patients received rehabilitation treatment at the Hubei Provincial Hospital of Traditional Chinese Medicine (Table 1).
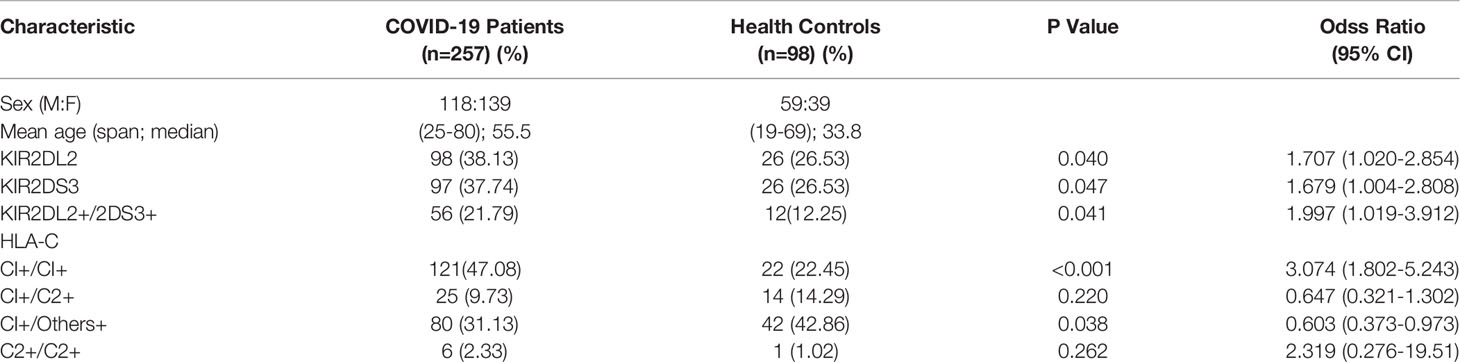
Table 1 Clinical characteristics and the frequencies of KIR2DL2, KIR2DS3, and HLA-C1 in COVID-19 patients and healthy persons.
KIR2DL2, KIR2DS3, and HLA-C1 Are Genetic Risk Factors for COVID-19 Infection
KIR gene frequency was compared between COVID-19 patients and healthy controls (Table 1). KIR2DL2 and KIR2DS3 were more highly frequent in infected than uninfected individuals (KIR2DL2: 38.13% vs 26.53%, respectively; OR=1.707, P=0.04; KIR2DS3: 37.74% vs 26.53%, OR=1.689, P=0.047). The frequency of KIR2DL2+/KIR2DS3+ was also higher in COVID-19 patients than healthy controls (21.79% vs 12.25%, respectively; OR=1.997, P=0.041, 95% CI=1.019–3.912). These data suggest that KIR2DL2 and KIR2DS3 may be genetic risk factors for SARS-CoV-2 infection.
KIR activity requires the participation of HLA ligands. The frequency of HLA-C1C1, the homozygous receptor for KIR2DL2 and KIR2DL3, was also higher in COVID-19 patients than in healthy controls (47.08% vs 22.45%, respectively; OR=3.074, P <0.001). It is suggested that the HLA-C1C1 gene pairing is a risk factor for COVID-19 (Table 1). Further analysis indicated that the frequency of KIR2DL2/HLA-C1+C1+ was higher in the COVID-19 patients than in the healthy controls (19.46% vs 10.2%, respectively; OR=2.126, P=0.038). These findings suggest that the KIR2DL2/HLA-C pairing is a genetic risk factor for COVID-19 (Table 2).
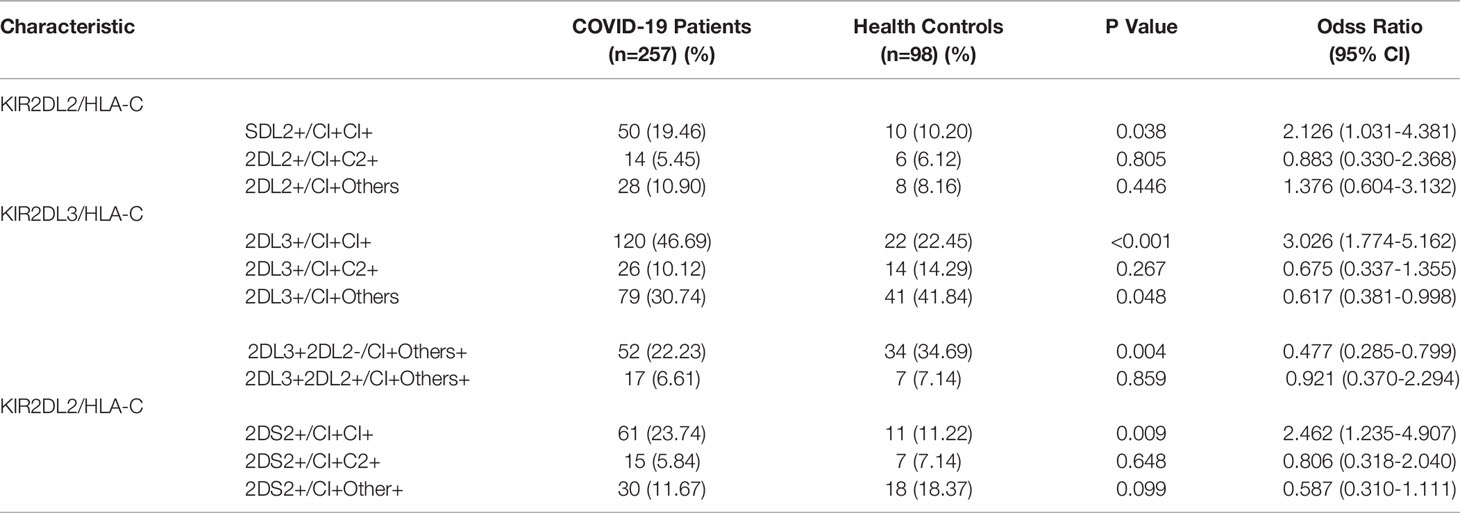
Table 2 Pairing analysis of the frequencies of KIR2DL2/2DL3/2DS2 and HLA-C in COVID-19 patients and healthy persons.
The KIR2DL3/HLA-C1 Pairing May be a Genetic Protective Factor Against COVID-19 Infection
Assessment of HLA-C distribution among the study subjects revealed a combination of HLA-C alleles in which one locus was C1 and the other locus was neither C1 nor C2. We termed this ‘Other’ or ‘O’ and named the combination HLA-C1+O+. The frequency of HLA-C1+O+ was lower among COVID-19 patients than healthy controls (31.13% vs 42.86%, respectively; OR = 0.603, P=0.038). Thus, it is possible that the HLA-C1+O+ combination is a genetic protective factor against SARS-CoV-2 infection (Table 2). Similarly, the KIR2DL3/HLA-C1+O+ combination was lower in the COVID-19 patients than in healthy individuals (30.74% vs 41.84%, respectively; OR =0.617, P= 0.048). To determine whether KIR2DL2 also plays a protection function, individuals with KIR2DL3/HLA-C1+O+ were divided into two groups based on their exist of KIR2DL2. COVID-19 patients were still less likely to carry KIR2DL3+KIR2DL2-/HLA-C1+O+ than healthy controls (22.23% vs 34.69%, respectively, OR =0.477, P=0.004), suggesting that this genetic combination is still protective in the absence of KIR2DL2 (Table 2). In contrast, the frequency of KIR2DL3+KIR2DL2+/HLA-C1+O+ was similar between COVID-19 patients and controls (6.61% vs 7.14%, respectively; OR =1.086, P=0.859). These results show that the protective effect of the KIR2DL3/HLA-C1 pairing is dependent on the absence of KIR2DL2.
KIR2DL2 Distribution Correlates Positively With the COVID-19 Caseload in Different Geographic Regions
To confirm the association between KIR2DL2 distribution and SARS-CoV-2 infection, we used public information platforms (https://www.worldometers.info/coronavirus/) to obtain the incidence of COVID-19 (Case/1 M pop) in different geographic regions as of May 10, 2021 (Figure 1).
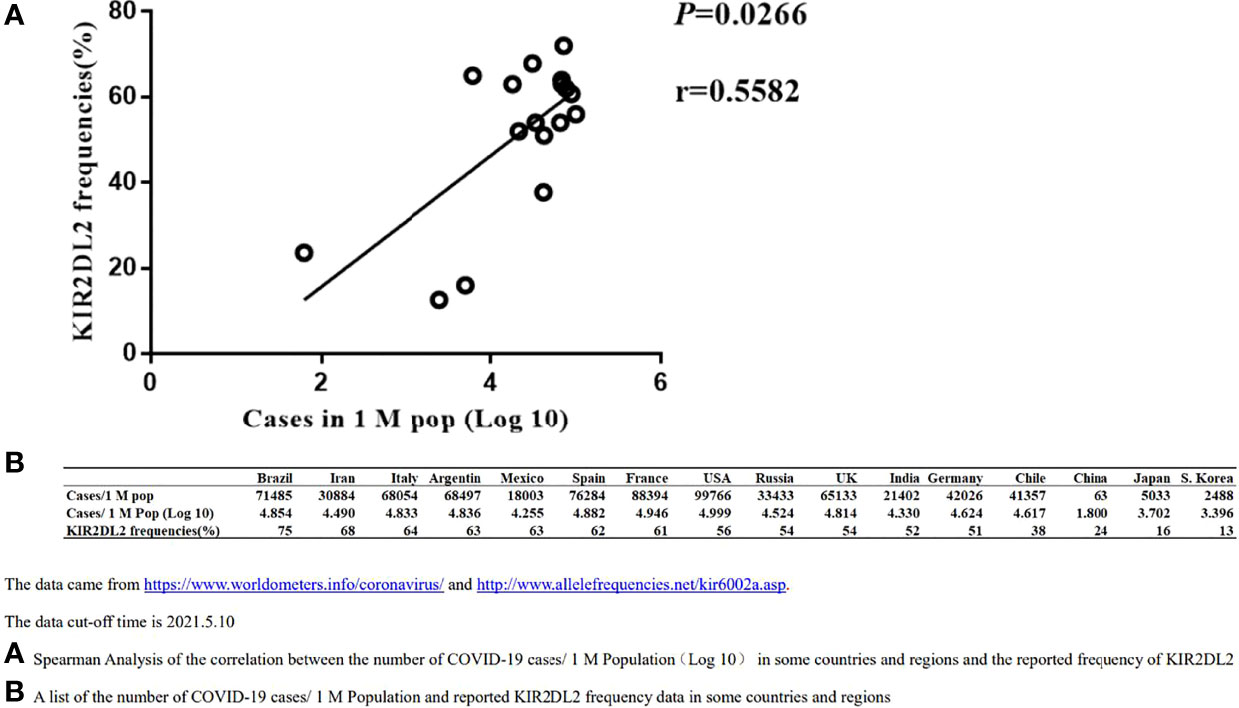
Figure 1 Spearman Correlation analysis between the distribution of KIR2DL2 and the occurrences of global COVID-19 cases. The data cut-off time is 2021.5.10. (A). Spearman Analysis of the correlation between the number of COVID-19 cases/1 M Population(Log 10) in some countries and regions and the reported frequency of KIR2DL2. (B). A list of the number of COVID-19 cases/1 M Population and reported KIR2DL2 frequency data in some countries and regions.
The following equation was used to calculate incidence:
Case/1 M pop = total number of COVID-19 cases/population of region*1,000,000
The frequency of KIR2DL2 in geographic regions with available public data was determined using relevant websites (http://www.allelefrequencies.net/kir6002a.asp), and the frequency of KIR2DL2 reported in each region was used for follow-up analysis. Spearman correlation analysis suggested that KIR2DL2 distribution was positively correlated with COVID-19 incidence (p=0.0266, r=0.5582) (Figure 1).
COVID-19 Severity May Not Correlate Directly With KIR Gene
To determine if KIR gene was associated with COVID-19 severity, the same combinations of KIR2DL2, KIR2DS3/HLA-C, and KIR2DL3/HLA-C were compared between patients with mild and severe disease. There were no significant differences in disease severity, suggesting that the KIR gene may not correlate with COVID-19 pathogenesis (Table 3).
Discussion
The occurrence of viral infections is shown to be closely related to an individual’s genetic background (22–24). NK cells are fast-acting innate immune cells that provide a critical first line of defense by killing infected cells and producing pro-inflammatory cytokines (25). The current study found that KIR2DL2 gene frequency was much higher in COVID-19 patients than in healthy controls. Interestingly, while there were no differences in KIR2DL3 in the initial analysis, this gene was found to have a protective effect when combined with HLA-C1+O+ in the absence of KIR2DL2. Indeed, the KIR2DL2/HLA-C1+C1+ pairing appeared more frequently in COVID-19 patients than in healthy individuals. We also analyzed current public data and found that the frequency of KIR2DL2 in the population correlated positively with the number of COVID-19 cases in particular geographic regions. These results suggest that people carrying the KIR2DL2/HLA-C1C1 gene may be at high risk for COVID-19. KIR2DL2 is known to inhibit NK activation, which may prevent the early clearance of SARS-CoV-2.
KIR plays an important role in aiding the immune response against virus-infected cells. KIR3DL1 is more highly frequent in HBV-infected individuals (26) and KIR2DS1 may reduce the risk of CMV infection after kidney transplantation (27). Our previous study found that the presence of KIR2DL2 inhibits the NK cell response to HCV-infected patients in the Chinese Han population. Indeed, KIR2DL2 is not only associated with the HCV infection rate but also with HCV clearance (28). These results suggest that KIRs play a role in the occurrence and persistence of infectious diseases.
The KIR2DL2 and KIR2DL3 cDNA sequences have been given different names, but genome analysis and population studies indicate that they may be alleles at the same locus (29), and share high homology. KIR2DL2 combines more strongly with HLA-C than KIR2DL3 (30) and may exert a more powerful inhibitory effect on NK cells that promotes survival of the virus. KIR expression on NK cells is largely random (31), so it is possible that some people carrying the KIR2DL2 gene may have more KIR2DL2+CD56dimNK cell subgroups that weaken the antiviral response and promote infection. KIR2DL3+ also plays inhibitory function but is weaker than KIR2DL2+, so KIR2DL3+ NK cells are more easily activated, which might inhibit the virus at the early stage of infection to prevent disease.
Our data implied that KIR/HLA-C is not related to the progression of COVID-19. One possible explanation is that CD4+ and CD8+ T lymphocytes are the main cells involved in fighting this disease, and contribute to the development of severe outcomes such as the “cytokine storm” (32). KIR is mainly expressed on the surface of CD56dim NK cells. This NK cell subtype is primarily involved in killing infected target cells, and cytokine secretion is limited (33). It is also possible that an association between KIR/HLA-C and COVID-19 progression was not observed because this study was primarily limited to the Chinese Han population. In a recent study of 424 COVID-19 positive patients (392 Saudi, 32 non-Saudi), the KIR2DS4 gene was associated with the highest risk of severe COVID-19 infection (OR 8.48, p= 0.0084) followed by KIR3DL1 (OR 7.61, p=0.0192) (34). The absence of KIR3DL1+HLA-Bw4+ and KIR3DL2+HLA-A3/11+ pairings were protective against COVID-19 progression in 200 hospitalized patients at the University of California San Francisco Medical Center during March–October 2020 (35). KIR genotypes vary widely across ethnic groups (36). All of these research suggested that KIR genes involved in COVID-19 progression, and different KIR gene might play different roles.
This study provides a correlation between the immune genetic background of some Han people and COVID-19. In particular, KIR2DL2/HLA-C1+C1+ may be a risk factor for SARS-CoV-2 infection in this population. KIR are a group of molecules that play an important role in regulating the differentiation, development, and activation of NK cells. The random expression of KIR genes among individuals may change the distribution of disease in different populations. The current study is primarily focused on gene frequency and disease. Further research will be required to characterize the mechanism of KIR expression, such as epigenetic regulation, in the occurrence and development of infectious diseases. This will help to inform our understanding of host–virus interactions.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The project was approved after discussion by the Ethics Committee of Hubei Province Academy of Traditional Chinese Medicine (HBZY2020-CO1-01). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
BS and SH jointly completed the experimental design and article writing. ZS and MX were mainly involved in the recruitment of COVID-19 survivors and the division of experimental cohorts. WN and PS were responsible for the collection and sorting of peripheral blood of COVID-19 survivors. HW, YH, and XL were mainly involved in PCR-SSP and result reading and recording. JQ focused on the quality control and analysis of results. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Wuhan Municipal Health Commission (No. EX20B02) and Jianghan University Funds (3015/06210035, 1010/08190006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
COVID-19, 2019 Coronavirus Disease; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; NK cells, Natural killer cells; HLA, Human leukocyte antigen; KIR, Killer cell immunoglobulin-like receptor.
References
1. Velavan TP, Meyer CG. The COVID-19 Epidemic. Trop Med Int Health: TM IH. (2020) 3):278–80. doi: 10.1111/tmi.13383
2. Kikkert M. Innate Immune Evasion by Human Respiratory RNA Viruses. J innate immunity. (2020) 12(1):4–20. doi: 10.1159/000503030
3. Bavel JJV, Baicker K, Boggio PS, Capraro V, Cichocka A, Cikara M, et al. Using Social and Behavioural Science to Support COVID-19 Pandemic Response. Nat Hum Behav (2020) 4(5):460–71. doi: 10.1038/s41562-020-0884-z
4. Qian GQ, Yang NB, Ma AHY, Wang LP, Li GX, Chen XQ, et al. COVID-19 Transmission Within a Family Cluster by Presymptomatic Carriers in China. Clin Infect Diseases. (2020) 71(15):861–2. doi: 10.1093/cid/ciaa316
5. Naemi FMA, Al-Adwani S, Al-Khatabi H, Al-Nazawi A. Frequency of HLA Alleles Among COVID-19 Infected Patients: Preliminary Data From Saudi Arabia. Virology (2021) 560:1–7. doi: 10.1016/j.virol.2021.04.011
6. Khor SS, Omae Y, Nishida N, Sugiyama M, Kinoshita N, Suzuki T, et al. HLA-A*11:01:01:01, HLA-C*12:02:02:01-HLA-B*52:01:02:02, Age and Sex Are Associated With Severity of Japanese COVID-19 With Respiratory Failure. Front Immunol (2021) 12:658570. doi: 10.3389/fimmu.2021.658570
7. Pisanti S, Deelen J, Gallina AM, Caputo M, Citro M, Abate M, et al. Correlation of the Two Most Frequent HLA Haplotypes in the Italian Population to the Differential Regional Incidence of Covid-19. J Trans Med (2020) 18(1):352. doi: 10.1186/s12967-020-02515-5
8. Merad M, Martin JC. Pathological Inflammation in Patients With COVID-19: A Key Role for Monocytes and Macrophages. Nat Rev Immunol (2020) 20(6):355–62. doi: 10.1038/s41577-020-0331-4
9. Qin W, Hu BZ, Zhang Z, Chen S, Li FJ, Zhu ZY, et al. Clinical Characteristics and Death Risk Factors of Severe COVID-19. Zhonghua Jie He He Hu Xi Za Zhi. (2020) 43(8):648–53. doi: 10.3760/cma.j.cn112147-20200320-00380
10. Jiang Y, Wei X, Guan J, Qin S, Wang Z, Lu H, et al. COVID-19 Pneumonia: CD8(+) T and NK Cells are Decreased in Number But Compensatory Increased in Cytotoxic Potential. Clin Immunol (2020), 218:108516. doi: 10.1016/j.clim.2020.108516
11. Colonna M, Nakajima H, Navarro F, Lopez-Botet M. A Novel Family of Ig-Like Receptors for HLA Class I Molecules That Modulate Function of Lymphoid and Myeloid Cells. J Leukocyte Biol (1999) 66(3):375–81. doi: 10.1002/jlb.66.3.375
12. Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity (2017) 47(5):820–33. doi: 10.1016/j.immuni.2017.10.008
13. Carrillo-Bustamante P, Kesmir C, de Boer RJ. The Evolution of Natural Killer Cell Receptors. Immunogenetics (2016) 68(1):3–18. doi: 10.1007/s00251-015-0869-7
14. Dogra P, Rancan C, Ma W, Toth M, Senda T, Carpenter DJ, et al. Tissue Determinants of Human NK Cell Development, Function, and Residence. Cell (2020) 180(4):749–63.e13. doi: 10.1016/j.cell.2020.01.022. Cited in: Pubmed
15. Pende D, Falco M, Vitale M, Cantoni C, Vitale C, Munari E, et al. Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front Immunol (2019) 10:1179. doi: 10.3389/fimmu.2019.01179
16. Nowak I, Wilczynska K, Wilczynski JR, Malinowski A, Radwan P, Radwan M, et al. LILRB and Their Ligands’ Genes as Potential Biomarkers in Recurrent Implantation Failure. Archivum Immunologiae Therapiae Experimentalis (2017) 65(5):391–9. doi: 10.1007/s00005-017-0474-6
17. Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK Cell Inhibitory Receptor Genes in Resolving Hepatitis C Virus Infection. Science (2004) 305(5685):872–4. doi: 10.1126/science.1097670
18. Hirayasu K, Ohashi J, Kashiwase K, Hananantachai H, Naka I, Ogawa A, et al. Significant Association of KIR2DL3-HLA-C1 Combination With Cerebral Malaria and Implications for Co-Evolution of KIR and HLA. PloS pathogens. (2012) 8(3):e1002565. doi: 10.1371/journal.ppat.1002565
19. Colucci F. The Role of KIR and HLA Interactions in Pregnancy Complications. Immunogenetics (2017) 69(8-9):557–65. doi: 10.1007/s00251-017-1003-9
20. Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-Like Receptor Haplotype Analysis by Gene Content: Evidence for Genomic Diversity With a Minimum of Six Basic Framework Haplotypes, Each With Multiple Subsets. J Immunol (2002) 169(9):5118–29. doi: 10.4049/jimmunol.169.9.5118
21. Bunce M, O’Neill CM, Barnardo MC, Krausa P, Browning MJ, Morris PJ, et al. Phototyping: Comprehensive DNA Typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR With 144 Primer Mixes Utilizing Sequence-Specific Primers (PCR-SSP). Tissue Antigens (1995) 46(5):355–67. doi: 10.1111/j.1399-0039.1995.tb03127.x
22. Lam JKP, Azzi T, Hui KF, Wong AMG, McHugh D, Caduff N, et al. Co-Infection of Cytomegalovirus and Epstein-Barr Virus Diminishes the Frequency of CD56(dim)NKG2A(+)KIR(-) NK Cells and Contributes to Suboptimal Control of EBV in Immunosuppressed Children With Post-Transplant Lymphoproliferative Disorder. Front Immunol (2020) 11:1231. doi: 10.3389/fimmu.2020.01231
23. Burek Kamenaric M, Ivkovic V, Kovacevic Vojtusek I, Zunec R. The Role of HLA and KIR Immunogenetics in BK Virus Infection After Kidney Transplantation. Viruses (2020) 12(12):1417. doi: 10.3390/v12121417.
24. Li Q, Liu S, Zhang S, Liu C, Sun M, Li C, et al. Human Leucocyte Antigen But Not KIR Alleles and Haplotypes Associated With Chronic HCV Infection in a Chinese Han Population. Int J immunogenetics (2019) 46(4):263–73. doi: 10.1111/iji.12425
25. Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural Killer Cells in Antiviral Defense: Function and Regulation by Innate Cytokines. Annu Rev Immunol (1999) 17:189–220. doi: 10.1146/annurev.immunol.17.1.189
26. Varbanova V, Popov G, Grigorova V, Petrova D, Naumova E, Mihaylova A. KIR/HLA Ligands Immunogenetics Markers Associated With Outcome of Hepatitis B Virus Infection in the Bulgarian Population. Biomed papers Med Faculty Univ Palacky Olomouc Czechoslovakia (2021) 165(3):270–6. doi: 10.5507/bp.2020.043
27. Farzamikia N, Hejazian SM, Haghi M, Hejazian SS, Zununi Vahed S, Ardalan M. Evaluation of Telomeric KIR Genes and Their Association With CMV Infection in Kidney Transplant Recipients. Immunogenetics (2022) 74(2):207–12. doi: 10.1007/s00251-021-01245-2
28. Hu S, Yuan F, Feng L, Zheng F, Gong F, Huang H, et al. KIR2DL2/C1 is a Risk Factor for Chronic Infection and Associated With Non-Response to PEG-IFN and RBV Combination Therapy in Hepatitis C Virus Genotype 1b Patients in China. Virologica Sinica (2018) 33(4):369–72. doi: 10.1007/s12250-018-0042-1
29. Uhrberg M, Parham P, Wernet P. Definition of Gene Content for Nine Common Group B Haplotypes of the Caucasoid Population: KIR Haplotypes Contain Between Seven and Eleven KIR Genes. Immunogenetics (2002) 54(4):221–9. doi: 10.1007/s00251-002-0463-7
30. Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic Polymorphism at Two Positions Distal to the Ligand-Binding Site Makes KIR2DL2 a Stronger Receptor for HLA-C Than KIR2DL3. J Immunol (2008) 180(6):3969–79. doi: 10.4049/jimmunol.180.6.3969
31. Parham P. MHC Class I Molecules and KIRs in Human History, Health and Survival. Nat Rev Immunol (2005) 5(3):201–14. doi: 10.1038/nri1570
32. Huang W, Berube J, McNamara M, Saksena S, Hartman M, Arshad T, et al. Lymphocyte Subset Counts in COVID-19 Patients: A Meta-Analysis. Cytometry Part A: J Int Soc Analytical Cytology (2020) 97(8):772–6. doi: 10.1002/cyto.a.24172
33. Cichocki F, Grzywacz B, Miller JS. Human NK Cell Development: One Road or Many? Front Immunol (2019) 10:2078. doi: 10.3389/fimmu.2019.02078
34. Hajeer A, Jawdat D, Massadeh S, Aljawini N, Abedalthagafi MS, Arabi YM, et al. Association of KIR Gene Polymorphisms With COVID-19 Disease. Clin Immunol (2022) 234:108911. doi: 10.1016/j.clim.2021.108911
35. Maruthamuthu S, Rajalingam K, Kaur N, Morvan MG, Soto J, Lee N, et al. Individualized Constellation of Killer Cell Immunoglobulin-Like Receptors and Cognate HLA Class I Ligands That Controls Natural Killer Cell Antiviral Immunity Predisposes COVID-19. Front Genet (2022) 13:845474. doi: 10.3389/fgene.2022.845474
Keywords: COVID-19, SARS-CoV-2, NK cells, HLA, KIR
Citation: Hu S, Shao Z, Ni W, Sun P, Qiao J, Wan H, Huang Y, Liu X, Zhai H, Xiao M and Sun B (2022) The KIR2DL2/HLA-C1C1 Gene Pairing Is Associated With an Increased Risk of SARS-CoV-2 Infection. Front. Immunol. 13:919110. doi: 10.3389/fimmu.2022.919110
Received: 13 April 2022; Accepted: 13 June 2022;
Published: 07 July 2022.
Edited by:
Nabila Jabrane-Ferrat, INSERM U1043 Centre de Physiopathologie de Toulouse Purpan, FranceReviewed by:
Jose Artur Chies, Federal University of Rio Grande do Sul, BrazilAntonio Giordano, Temple University, United States
Copyright © 2022 Hu, Shao, Ni, Sun, Qiao, Wan, Huang, Liu, Zhai, Xiao and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binlian Sun, binlian17@jhun.edu.cn; Mingzhong Xiao, 309452513@qq.com
†These authors have contributed equally to this work
 Song Hu
Song Hu Zuoyu Shao2,3†
Zuoyu Shao2,3† Mingzhong Xiao
Mingzhong Xiao Binlian Sun
Binlian Sun