SARS-CoV-2: Repurposed Drugs and Novel Therapeutic Approaches—Insights into Chemical Structure—Biological Activity and Toxicological Screening
Abstract
:1. Introduction
2. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS–CoV-2)—A Brief Portrait
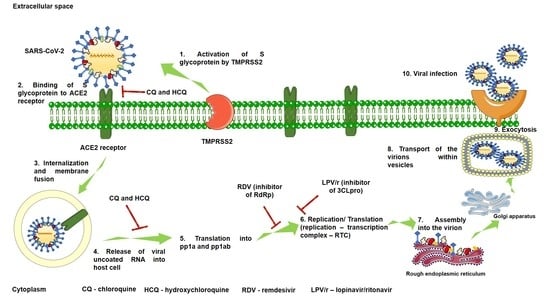
2.1. Mechanism of SARS-CoV-2 Viral Infection
2.2. Clinical Impact of SARS-CoV-2
3. Drugs Recommended in COVID-19 Therapeutic Guidelines
3.1. Remdesivir—RDV (GS-5734)
- The duration of RDV treatment should be settled at 5 days (200 mg day 1 followed by 100 mg/daily for the following 4 days) in severe COVID-19 cases—on the basis of the results obtained within SIMPLE trial 1, this measure represents a major step in reducing the risk of drug–drug interactions/side-effects/complications/aggravation of patient status;
- As the RDV directly-linked adverse effect is hepatic injury, a possible preventive approach would consist of the co-administration of hepatoprotective medication (for example, essentials phospholipids or albumin solution—it should be a patient-dependent decision after analyzing the risk/benefit ratio) during the RDV treatment (in severely ill COVID-19 patients, liver dysfunction was reported, but until now, it is unclear what causes it—the medication administered or the SARS-CoV-2 infection, thus liver protection would do no harm);
- As RDV is predominantly eliminated through urine (mostly as metabolites), the renal function should be monitored during the treatment and, in the case of renal impairment, the treatment should be stopped
- Co-administration of other anti-COVID-19 drugs (lopinavir/ritonavir, chloroquine, hydroxychloroquine, interferon) is possible because no drug–drug interactions were reported; surveillance is recommended
- Co-administration of antidiabetics and cardiovascular medication (anti-hypertensives, anticoagulants, beta blockers, and so on) is possible because no drug–drug interactions were reported; surveillance is recommended
- Co-administration of RDV with other hepatotoxic drugs is not recommended
- To remedy/to prevent the mild adverse reactions reported in the clinical trials, such as nausea, diarrhea, hyperlipidemia, and so on, symptomatic medication could be administered during RDV treatment because no interactions were described between these classes of drugs and RDV; strict surveillance is recommended
- To follow the updates on other clinical trials that tested RDV.
3.2. Chloroquine and Hydroxychloroquine
3.3. Lopinavir/Ritonavir
4. Potential Promising Antiviral Agents
4.1. Oseltamivir
4.2. Ribavirin
4.3. Arbidol Hydrochloride (Umifenovir)
4.4. Favipiravir
4.5. Betulinic Acid
4.6. Anti-Inflammatory Compounds
4.7. Immunotherapy
4.8. Anticoagulant Therapy
5. Closing Remarks and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Khan, S.; Siddique, R.; Shereen, M.A.; Ali, A.; Liu, J.; Bai, Q.; Bashir, N.; Xue, M. Emergence of a Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2: Biology and Therapeutic Options. J. Clin. Microbiol. 2020, 58, e00187-20. [Google Scholar] [CrossRef] [Green Version]
- Sohrabi, C.; Alsafi, Z.; O’Neill, N.; Khan, M.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, R. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 2020, 76, 71–76. [Google Scholar] [CrossRef]
- Lai, C.C.; Liu, Y.H.; Wang, C.Y.; Wang, Y.H.; Hsueh, S.C.; Yen, M.Y.; Ko, W.C.; Hsueh, P.R. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. J. Microbiol. Immunol. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Home Page—WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 25 April 2020).
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- COVID-19 Map. Available online: https://news.google.com/covid19/map?hl=en-US&gl=US&ceid=US:en (accessed on 4 May 2020).
- ECDC. COVID-19 Situation Update Worldwide, as of 10 May 2020. Available online: https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases (accessed on 11 May 2020).
- WHO. Home Page—Coronavirus Disease (COVID-19) Situation Report—105. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200504-covid-19-sitrep-105.pdf?sfvrsn=4cdda8af_2 (accessed on 5 May 2020).
- Wu, D.; Wu, T.; Liu, Q.; Yang, Z. The SARS-CoV-2 Outbreak: What We Know. Int. J. Infect. Dis. 2020, 94, 44–48. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Holmes, E.C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell 2020, 181, 223–227. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Shanmugaraj, B.; Siriwattananon, K.; Wangkanont, K.; Phoolcharoen, W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19). Asian Pac. J. Allergy Immunol. 2020, 38, 10–18. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1. Ref. Modul. Life Sci. 2020. [Google Scholar] [CrossRef]
- Ashour, H.M.; Elkhatib, W.F.; Rahman, M.M.; Elshabrawy, H.A. Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks. Pathogens 2020, 9, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kannan, S.; Shaik Syed Ali, P.; Sheeza, A.; Hemalatha, K. COVID-19 (Novel Coronavirus 2019)—Recent Trends. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2006–2011. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Lu, X.; Xu, C.; Sun, W.; Pan, B. Understanding of COVID-19 Based on Current Evidence. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- NCBI. GenBank. Available online: https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/ (accessed on 18 June 2020).
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic Characterization of a Novel SARS-CoV-2. Gene Rep. 2020, 19, 100682. [Google Scholar] [CrossRef]
- Penarrubia, A.L.; Ruiz, M.; Porco, R.; Rao, S.N.; Juanola-Falgarona, M.; Manissero, D.; López-Fontanals, M.; Pareja, J. Multiple Assays in a Real-Time RT-PCR SARS-CoV-2 Panel Can Mitigate the Risk of Loss of Sensitivity by New Genomic Variants during the COVID-19 Outbreak. Int. J. Infect. Dis. Off. Publ. Int. Soc. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Fu, Y.; Cheng, Y.; Wu, Y. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol. Sin. 2020. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Qiao, S.; Zhang, G. Analysis of Angiotensin-Converting Enzyme 2 (ACE2) From Different Species Sheds Some Light on Cross-Species Receptor Usage of a Novel Coronavirus 2019-nCoV. J. Infect. 2020, 80, 469–496. [Google Scholar] [CrossRef] [Green Version]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The Origin, Transmission and Clinical Therapies on Coronavirus Disease 2019 (COVID-19) Outbreak—An Update on the Status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Neurath, M.F. Covid-19 and Immunomodulation in IBD. Gut 2020. [Google Scholar] [CrossRef]
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.C.; Deng, Q.X.; Dai, S.X. Remdesivir for Severe Acute Respiratory Syndrome Coronavirus 2 Causing COVID-19: An Evaluation of the Evidence. Travel Med. Infect. Dis. 2020, 101647. [Google Scholar] [CrossRef]
- Nicola, M.; O’Neill, N.; Sohrabi, C.; Khan, M.; Agha, M.; Agha, R. Evidence based management guideline for the COVID-19 pandemic—Review article. Int. J. Surg. 2020, 77, 206–216. [Google Scholar] [CrossRef]
- Tian, Y.; Rong, L.; Nian, W.; He, Y. Review Article: Gastrointestinal Features in COVID-19 and the Possibility of Faecal Transmission. Aliment. Pharmacol. Ther. 2020, 51, 843–851. [Google Scholar] [CrossRef]
- Kluge, S.; Janssens, U.; Welte, T.; Weber-Carstens, S.; Marx, G.; Karagiannidis, C. German Recommendations for Critically Ill Patients with COVID-19. Med. Klin. Intensivmed. Notfmed. 2020. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [Green Version]
- Kuba, K.; Imai, Y.; Ohto-Nakanishi, T.; Penninger, J.M. Trilogy of ACE2: A Peptidase in the Renin-Angiotensin System, a SARS Receptor, and a Partner for Amino Acid Transporters. Pharmacol. Ther. 2010, 128, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Docea, A.O.; Tsatsakis, A.; Albulescu, D.; Cristea, O.; Zlatian, O.; Vinceti, M.; Moschos, S.A.; Tsoukalas, D.; Goumenou, M.; Drakoulis, N.; et al. A new threat from an old enemy: Re-emergence of coronavirus (Review). Int. J. Mol. Med. 2020, 45, 1631–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, G.; Zheng, K.I.; Yan, Q.Q.; Rios, R.S.; Targher, G.; Byrne, C.D.; Poucke, S.V.; Liu, W.Y.; Zheng, M.H. COVID-19 and Liver Dysfunction: Current Insights and Emergent Therapeutic Strategies. J. Clin. Transl. Hepatol. 2020, 8, 18–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bikdeli, B.; Madhavan, M.V.; Jimenez, D.; Chuich, T.; Dreyfus, I.; Driggin, E.; Nigoghossian, C.; Ageno, W.; Madjid, M.; Guo, Y.; et al. COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-up. J. Am. Coll. Cardiol. 2020. [Google Scholar] [CrossRef]
- Meng, X.; Deng, Y.; Dai, Z.; Meng, Z. COVID-19 and Anosmia: A Review Based on Up-To-Date Knowledge. Am. J. Otolaryngol. 2020, 41, 102581. [Google Scholar] [CrossRef]
- Dawson, P.; Rabold, E.M.; Laws, R.L.; Conners, E.E.; Gharpure, R.; Yin, S.; Buono, S.A.; Dasu, T.; Bhattacharyya, S.; Westergaard, R.P.; et al. Loss of Taste and Smell as Distinguishing Symptoms of COVID-19. Clin. Infect. Dis. 2020, 21. [Google Scholar] [CrossRef]
- Li, J.; Fan, J.G. Characteristics and Mechanism of Liver Injury in 2019 Coronavirus Disease. J. Clin. Transl. Hepatol. 2020, 8, 13–17. [Google Scholar] [CrossRef] [Green Version]
- Hong, E.H.; Song, J.H.; Kang, K.B.; Sung, S.H.; Ko, H.J.; Yang, H. Anti-Influenza Activity of Betulinic Acid From Zizyphus Jujuba on Influenza A/PR/8 Virus. Biomol. Ther. 2015, 23, 345–349. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, R. Antibody-Dependent Enhancement of Viral Infections. Dyn. Immune Act. Viral Dis. 2019, 5, 9–41. [Google Scholar] [CrossRef] [Green Version]
- Davies, N.G.; Klepac, P.; Liu, Y.; Prem, K.; Jit, M.; CMMID COVID-19 Working Group; Eggo, R.M. Age-dependent Effects in the Transmission and Control of COVID-19 Epidemics. Nat. Med. 2020. [Google Scholar] [CrossRef]
- Xie, Z. Pay attention to SARS-CoV-2 infection in children. Pediatric Investig. 2020, 4, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Liguoro, I.; Pilotto, C.; Bonanni, M.; Ferrari, M.E.; Pusiol, A.; Nocerino, A.; Vidal, E.; Cogo, P. SARS-COV-2 infection in children and newborns: A systematic review. Eur. J. Pediatr. 2020. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhou, Q.; Li, Y.; Garner, L.V.; Watkins, S.P.; Carter, L.J.; Smoot, J.; Gregg, A.C.; Daniels, A.D.; Jervey, S.; et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 2020, 6, 315–331. [Google Scholar] [CrossRef] [PubMed]
- WHO. Solidarity Clinical Trial for COVID-19 Treatments. Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---18-march-2020 (accessed on 25 April 2020).
- WHO. Public Statement for Collaboration on COVID-19 Vaccine Development. Available online: https://www.who.int/news-room/detail/13-04-2020-public-statement-for-collaboration-on-covid-19-vaccine-development (accessed on 25 April 2020).
- NIH U.S National Library of Medicine. Home—ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/results?cond=COVID-19 (accessed on 11 May 2020).
- WHO. WHO | Welcome to the WHO ICTRP. Available online: https://www.who.int/ictrp/en/ (accessed on 30 April 2020).
- Van Ierssel, S.; Dauby, N.; Bottieau, E.; Huits, R.; The Belgium Task Force. Interim Clinical Guidance for Adults with Suspected or Confirmed COVID-19 in Belgium 06 May 2020, Version 8. 2020.
- Van Ierssel, S.; Dauby, N.; Bottieau, E.; Huits, R.; The Belgium Task Force. Interim Clinical Guidance for Adults with Suspected or Confirmed COVID-19 in Belgium 06 May 2020, Version 5. 2020.
- Lombardy Section Italian Society Infectious and Tropical Diseases. Vademecum for the treatment of people with COVID-19. Edition 2.0, 13 March 2020. Infez. Med. 2020, 28, 143–152. [Google Scholar]
- Kupferschmidt, K.; Cohen, J. Race to Find COVID-19 Treatments Accelerates. Science 2020, 367, 1412–1413. [Google Scholar] [CrossRef] [Green Version]
- Gilead. Gilead Announces Results from Phase 3 Trial of Investigational Antiviral Remdesivir in Patients with Severe COVID-19. Available online: https://www.gilead.com/news-and-press/press-room/press-releases/2020/4/gilead-announces-results-from-phase-3-trial-of-investigational-antiviral-remdesivir-in-patients-with-severe-covid-19 (accessed on 30 April 2020).
- A NIH NIAID. NIH Clinical Trial Shows Remdesivir Accelerates Recovery from Advanced COVID-19 | NIH: National Institute of Allergy and Infectious Diseases. Available online: https://www.niaid.nih.gov/news-events/nih-clinical-trial-shows-remdesivir-accelerates-recovery-advanced-covid-19 (accessed on 30 April 2020).
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9, e00221-18. [Google Scholar] [CrossRef] [Green Version]
- Ko, W.C.; Rolain, J.M.; Lee, N.Y.; Chen, P.L.; Huang, C.T.; Lee, P.I.; Hsueh, P.R. Arguments in Favour of Remdesivir for Treating SARS-CoV-2 Infections. Int. J. Antimicrob. Agents 2020, 55, 105933. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and Chloroquine Effectively Inhibit the Recently Emerged Novel Coronavirus (2019-nCoV) in Vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Götte, M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses 2019, 11, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. wissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Huang, S.; Zheng, F.; Dai, Y. Controversial treatments: An updated understanding of the coronavirus disease 2019. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amirian, E.S.; Levy, J.K. Current Knowledge about the Antivirals Remdesivir (GS-5734) and GS-441524 as Therapeutic Options for Coronaviruses. One Health 2020, 9, 100128. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.K.; Jordan, R.; Arvey, A.; Sudhamsu, J.; Shrivastava-Ranjan, P.; Hotard, A.L.; Flint, M.; McMullan, L.K.; Siegel, D.; Clarke, M.O.; et al. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017, 7, 43395. [Google Scholar] [CrossRef] [PubMed]
- Shannon, A.; Le, N.T.; Selisko, B.; Eydoux, C.; Alvarez, K.; Guillemot, J.C.; Decroly, E.; Peersen, O.; Ferron, F.; Canard, B. Remdesivir and SARS-CoV-2: Structural Requirements at Both nsp12 RdRp and nsp14 Exonuclease Active-Sites. Antiviral Res. 2020, 178, 104793. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 95, 6785–6797. [Google Scholar] [CrossRef] [Green Version]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural Basis for Inhibition of the RNA-dependent RNA Polymerase From SARS-CoV-2 by Remdesivir. Science 2020. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, R. Binding mechanism of remdesivir to SARS-CoV-2 RNA dependent RNA polymerase. Published online ahead of print: 17 March 2020. [CrossRef]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.X.; et al. Compassionate Use of Remdesivir for Patients With Severe Covid-19. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, D.; Du, G.; Du, R.; Zhao, J.; Jin, Y.; Fu, S.; Gao, L.; Cheng, Z.; Lu, Q.; et al. Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020. [Google Scholar] [CrossRef]
- EMA. Conditions of Use, Conditions for Distribution and Patients Targeted and Conditions for Safety Monitoring Adressed to Member States for Remdesivir Available for Compassionate Use. Available online: https://www.ema.europa.eu/en/documents/other/conditions-use-conditions-distribution-patients-targeted-conditions-safety-monitoring-adressed_en-0.pdf (accessed on 21 November 2013).
- Gilead. Gilead Announces Results from Phase 3 Trial of Remdesivir in Patients with Moderate COVID-19. Available online: https://www.gilead.com/news-and-press/press-room/press-releases/2020/6/gilead-announces-results-from-phase-3-trial-of-remdesivir-in-patients-with-moderate-covid-19 (accessed on 19 June 2020).
- Gilead. Gilead Announces Approval of Veklury® (Remdesivir) in Japan for Patients with Severe COVID-19. Available online: https://www.gilead.com/news-and-press/press-room/press-releases/2020/5/gilead-announces-approval-of-veklury-remdesivir-in-japan-for-patients-with-severe-covid19 (accessed on 19 June 2020).
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of Covid-19—Preliminary Report. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- EMA. EMA Starts Rolling Review of Remdesivir for COVID-19. Available online: https://www.ema.europa.eu/en/news/ema-starts-rolling-review-remdesivir-covid-19 (accessed on 30 June 2020).
- U.S. Food and Drug Administration. Remdesivir EUA Letter of Authorization; Gilead Sciences, Inc.: Foster City, CA, USA, 2020.
- EMA. Summary on Compassionate Use Remdesivir Gilead. Available online: https://www.ema.europa.eu/en/documents/other/summary-compassionate-use-remdesivir-gilead_en.pdf (accessed on 25 April 2020).
- The University of Liverpool. COVID-19 Drug Interactions, 2020 Detailed Recommendations for Interactions with Experimental COVID-19 Therapies. Available online: https://www.covid19-druginteractions.org/ (accessed on 25 April 2020).
- Hu, T.Y.; Frieman, M.; Wolfram, J. Insights from Nanomedicine into Chloroquine Efficacy Against COVID-19. Nat. Nanotechnol. 2020, 15, 247–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Ye, F.; Zhang, M.; Cui, C.; Huang, B.; Niu, P.; Liu, X.; Zhao, L.; Dong, E.; Song, C.; et al. Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Cao, R.; Xu, M.; Wang, X.; Zhang, H.; Hu, H.; Li, Y.; Hu, Z.; Zhong, W.; Wang, M. Hydroxychloroquine, a Less Toxic Derivative of Chloroquine, Is Effective in Inhibiting SARS-CoV-2 Infection in Vitro. Cell Discov. 2020, 6, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, A.K.; Kumar, A.; Tiwari, G.; Kumar, R.; Misra, N. In Silico Investigations on the Potential Inhibitors for COVID-19 Protease. Published Online Ahead of Print: 23 March 2020. Available online: http://arxiv.org/abs/2003.10642 (accessed on 25 April 2020).
- Colson, P.; Rolain, J.M.; Lagier, J.C.; Brouqui, P.; Raoult, D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105932. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.J.; Bergeron, E.; Benjannet, S.; Erickson, B.R.; Rollin, P.E.; Ksiazek, T.G.; Seidah, N.G.; Nichol, S.T. Chloroquine Is a Potent Inhibitor of SARS Coronavirus Infection and Spread. Virol. J. 2005, 2, 69. [Google Scholar] [CrossRef] [Green Version]
- Savarino, A.; Di Trani, L.; Donatelli, I.; Cauda, R.; Cassone, A. New Insights into the Antiviral Effects of Chloroquine. Lancet Infect. Dis. 2006, 6, 67–69. [Google Scholar] [CrossRef]
- Mingo, R.M.; Simmons, J.A.; Shoemaker, C.J.; Nelson, E.A.; Schornberg, K.L.; D’Souza, R.S.; Casanova, J.E.; White, J.M. Ebola Virus and Severe Acute Respiratory Syndrome Coronavirus Display Late Cell Entry Kinetics: Evidence That Transport to NPC1+ Endolysosomes Is a Rate-Defining Step. J. Virol. 2015, 89, 2931–2943. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of Therapeutic Targets for SARS-CoV-2 and Discovery of Potential Drugs by Computational Methods. Acta Pharm. Sin. B 2020. [Google Scholar] [CrossRef]
- PDR. Chloroquine Phosphate—Drud Summary. Available online: https://www.pdr.net/drug-summary/Chloroquine-Phosphate-chloroquine-phosphate-3418.2640 (accessed on 30 April 2020).
- Chowdhury, M.S.; Rathod, J.; Gernsheimer, J. A Rapid Systematic Review of Clinical Trials Utilizing Chloroquine and Hydroxychloroquine as a Treatment for COVID-19. Acad. Emerg. Med. 2020. [Google Scholar] [CrossRef]
- Chen, J.; Liu, D.; Liu, L.; Liu, P.; Xu, Q.; Xia, L.; Ling, Y.; Huang, D.; Song, S.; Zhang, D.; et al. A Pilot Study of Hydroxychloroquine in Treatment of Patients with Moderate COVID-19. Zhejiang Da Xue Xue Bao Yi Xue Ban 2020, 49, 215–219. [Google Scholar]
- Gautret, P.; Lagier, J.C.; Parola, P.; Hoang, V.T.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; et al. Hydroxychloroquine and Azithromycin as a Treatment of COVID-19: Results of an Open-Label Non-Randomized Clinical Trial. Int. J. Antimicrob. Agents 2020, 105949. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Tian, Z.; Yang, X. Breakthrough: Chloroquine Phosphate Has Shown Apparent Efficacy in Treatment of COVID-19 Associated Pneumonia in Clinical Studies. Biosci. Trends 2020, 14, 72–73. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.; Dai, S.M.; Tong, Q. COVID-19: A Recommendation to Examine the Effect of Hydroxychloroquine in Preventing Infection and Progression. J. Antimicrob. Chemother. 2020. [Google Scholar] [CrossRef]
- Meo, S.A.; Klonoff, D.C.; Akram, J. Efficacy of Chloroquine and Hydroxychloroquine in the Treatment of COVID-19. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4539–4547. [Google Scholar] [CrossRef] [PubMed]
- Gendelman, O.; Amital, H.; Bragazzi, N.L.; Watad, A.; Chodick, G. Continuous Hydroxychloroquine or Colchicine Therapy Does Not Prevent Infection with SARS-CoV-2: Insights from a Large Healthcare Database Analysis. Autoimmun. Rev. 2020, 102566. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, Y.; Fan, X.; Wang, X.; Han, Q.; Liu, Z. Advances in the Use of Chloroquine and Hydroxychloroquine for the Treatment of COVID-19. Postgrad. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.M.; Archibald, L.K.; Shukla, A.W.; Mehta, H.J.; Cherabuddi, K. Chloroquine and Hydroxychloroquine in the Context of COVID-19. Drugs Context 2020, 9. [Google Scholar] [CrossRef]
- Van den Eynde, J.J. COVID-19: An Update about the Discovery Clinical Trial. Pharmaceuticals 2020, 13, 98. [Google Scholar] [CrossRef]
- Pastick, K.A.; Okafor, E.C.; Wang, F.; Lofgren, S.M.; Skipper, C.P.; Nicol, M.R.; Pullen, M.F.; Rajasingham, R.; McDonald, E.G.; Lee, T.C.; et al. Review: Hydroxychloroquine and Chloroquine for Treatment of SARS-CoV-2 (COVID-19). Open Forum Infect. Dis. 2020, 7. [Google Scholar] [CrossRef] [Green Version]
- Meyerowitz, E.A.; Vannier, A.; Friesen, M.; Schoenfeld, S.; Gelfand, J.A.; Callahan, M.V.; Kim, A.Y.; Reeves, P.M.; Poznansky, M.C. Rethinking the Role of Hydroxychloroquine in the Treatment of COVID-19. FASEB J. 2020, 34, 6027–6037. [Google Scholar] [CrossRef]
- Ruamviboonsuk, P.; Lai, T.; Chang, A.; Lai, C.C.; Mieler, W.F.; Lam, D.; Asia-Pacific Vitreo-Retina Society. Chloroquine and Hydroxychloroquine Retinal Toxicity Consideration in the Treatment of COVID-19. Asia Pac. J. Ophthalmol. 2020, 9, 85–87. [Google Scholar] [CrossRef]
- Van den Broek, M.; Möhlmann, J.E.; Abeln, B.; Liebregts, M.; van Dijk, V.F.; van de Garde, E. Chloroquine-induced QTc Prolongation in COVID-19 Patients. Neth. Heart J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Szekely, Y.; Lichter, Y.; Shrkihe, B.A.; Bruck, H.; Oster, H.S.; Viskin, S. Chloroquine-induced torsades de pointes in a patient with coronavirus disease 2019. Heart Rhythm 2020, 30420–30423. [Google Scholar] [CrossRef] [PubMed]
- EMA. COVID-19: Chloroquine and Hydroxychloroquine Only to be Used in Clinical Trials or Emergency Use Programmes. Available online: https://www.ema.europa.eu/en/documents/press-release/covid-19-chloroquine-hydroxychloroquine-only-be-used-clinical-trials-emergency-use-programmes_en.pdf (accessed on 1 April 2020).
- WHO. Solidarity Clinical Trial for COVID-19 Treatments. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments (accessed on 19 June 2020).
- Muralidharan, N.; Sakthivel, R.; Velmurugan, D.; Gromiha, M.M. Computational Studies of Drug Repurposing and Synergism of Lopinavir, Oseltamivir and Ritonavir Binding with SARS-CoV-2 Protease against COVID-19. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.M.; Cheng, V.C.; Hung, I.F.; Wong, M.M.; Chan, K.H.; Chan, K.S.; Kao, R.Y.; Poon, L.L.; Wong, C.L.; Guan, Y.; et al. Role of Lopinavir/Ritonavir in the Treatment of SARS: Initial Virological and Clinical Findings. Thorax 2004, 59, 252–256. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.Y.; Chou, C.Y.; Chang, H.C.; Hsu, W.C.; Chang, G.G. Correlation between Dissociation and Catalysis of SARS-CoV Main Protease. Arch Biochem. Biophys. 2008, 472, 34–42. [Google Scholar] [CrossRef]
- Dayer, M.R.; Taleb-Gassabi, S.; Dayer, M.S. Lopinavir; a potent drug against coronavirus infection: Insight from molecular docking study. Arch. Clin. Infect. Dis. 2017, 12. [Google Scholar] [CrossRef] [Green Version]
- Tu, Y.F.; Chien, C.S.; Yarmishyn, A.A.; Lin, Y.Y.; Luo, Y.H.; Lin, Y.T.; Lai, W.Y.; Yang, D.M.; Chou, S.J.; Yang, Y.P.; et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int. J. Mol. Sci. 2020, 21, 2657. [Google Scholar] [CrossRef] [Green Version]
- ESICM. Surviving Sepsis Campaign Rapid Guidelines on the Management of Critically Ill Adults with Coronavirus Disease 2019. Available online: https://www.esicm.org/ssc-covid19-guidelines/ (accessed on 1 May 2020).
- EMA. Kaletra, Product Sheet 2020. Available online: https://www.ema.europa.eu/en/documents/product-information/kaletra-epar-product-information_en.pdf (accessed on 25 April 2020).
- Sun, J.; Deng, X.; Chen, X.; Huang, J.; Huang, S.; Li, Y.; Feng, J.; Liu, J.; He, G. Incidence of Adverse Drug Reactions in COVID-19 Patients in China: An Active Monitoring Study by Hospital Pharmacovigilance System. Clin. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef]
- IDSA. Infectious Diseases Society of America Guidelines on the Treatment and Management of Patients with COVID-19. Available online: https://www.idsociety.org/practice-guideline/covid-19-guideline-treatment-and-management/ (accessed on 25 April 2020).
- Gori, T.; Lelieveld, J.; Münzel, T. Perspective: Cardiovascular Disease and the Covid-19 Pandemic. Basic Res. Cardiol. 2020, 115, 32. [Google Scholar] [CrossRef]
- Bassetti, M.; Giacobbe, D.R.; Aliberti, S.; Barisione, E.; Centanni, S.; De Rosa, F.G.; Di Marco, F.; Gori, A.; Granata, G.; Mikulska, M.; et al. Balancing Evidence and Frontline Experience in the Early Phases of the COVID-19 Pandemic: Current Position of the Italian Society of Anti-infective Therapy (SITA) and the Italian Society of Pulmonology (SIP). Clin. Microbiol. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Naksuk, N.; Lazar, S.; Peeraphatdit, T.B. Cardiac Safety of Off-Label COVID-19 Drug Therapy: A Review and Proposed Monitoring Protocol. Eur. Heart J. Acute Cardiovasc. Care 2020. [Google Scholar] [CrossRef] [PubMed]
- Nitulescu, G.M.; Paunescu, H.; Moschos, S.A.; Petrakis, D.; Nitulescu, G.; Ion, G.N.D.; Spnadidos, D.A.; Nikolouzakis, T.K.; Drakoulis, N.; Tsatsakis, A. Comprehensive Analysis of Drugs to Treat SARS-CoV-2 Infection: Mechanistic Insights Into Current COVID-19 Therapies (Review). Int. J. Mol. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lew, W.; Chen, X.; Kim, C.U. Discovery and Development of GS 4104 (Oseltamivir): An Orally Active Influenza Neuraminidase Inhibitor. Curr. Med. Chem. 2000, 7, 663–672. [Google Scholar] [CrossRef] [Green Version]
- Davies, B.E. Pharmacokinetics of Oseltamivir: An Oral Antiviral for the Treatment and Prophylaxis of Influenza in Diverse Populations. J. Antimicrob. Chemother. 2010, 65. [Google Scholar] [CrossRef] [Green Version]
- Thomas, E.; Ghany, M.G.; Liang, T.J. Review the application and mechanism of action of ribavirin in therapy of hepatitis C. Antivir. Chem. Chemother. 2012, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020, 248, 117477. [Google Scholar] [CrossRef]
- Crotty, S.; Cameron, C.; Andino, R. Ribavirin’s Antiviral Mechanism of Action: Lethal Mutagenesis? J. Mol. Med. 2002, 80, 86–95. [Google Scholar] [CrossRef]
- Elfiky, A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592. [Google Scholar] [CrossRef]
- Mousavizadeh, L.; Ghasemi, S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020. [Google Scholar] [CrossRef]
- Dong, L.; Hu, S.; Gao, J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov. Ther. 2020, 14, 58–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.X.; Chen, X.P. Favipiravir: Pharmacokinetics and Concerns about Clinical Trials for 2019-NCoV Infection. Clin. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Q.; Yang, M.; Liu, D.; Chen, J.; Shu, D.; Xia, J.; Liao, X.; Gu, Y.; Cai, Q.; Yang, Y.; et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, Y.; Huang, J.; Yin, P.; Cheng, Z.; Wu, J.; Chen, S.; Zhang, Y.; Chen, B.; Lu, M.; et al. Favipiravir versus Arbidol for COVID-19: A Randomized Clinical Trial. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Russian Ministry of Health. Government Registry of Medicines, Medicine Registration License ЛП-006225. Available online: https://grls.rosminzdrav.ru/Grls_View_v2.aspx?routingGuid=38ce634a-8cb4-43ee-9283-5406335095ee&t= (accessed on 18 June 2020).
- Khaerunnisa, S.; Kurniawan, H.; Awaluddin, R.; Suhartati, S. Potential Inhibitor of COVID-19 Main Protease (M pro) from Several Medicinal Plant Compounds by Molecular Docking Study. Preprints 2020. [Google Scholar] [CrossRef] [Green Version]
- Ríos, J.L.; Máñez, S. New Pharmacological Opportunities for Betulinic Acid. Planta Med. 2018, 84, 8–19. [Google Scholar] [CrossRef] [Green Version]
- Phillips, J.; Phillips, I.; Enya, B.; Zhao, H.; Nitta, T. Effect of Betulinic Acid and Its Ionic Derivatives on M-MuLV Replication. Biochem. Biophys. Res. Commun. 2018, 500, 365–369. [Google Scholar] [CrossRef]
- Wen, C.C.; Kuo, Y.H.; Jan, J.T.; Liang, P.H.; Wang, S.Y.; Liu, H.G.; Lee, C.K.; Chang, S.T.; Kuo, C.J.; Lee, S.S.; et al. Specific Plant Terpenoids and Lignoids Possess Potent Antiviral Activities Against Severe Acute Respiratory Syndrome Coronavirus. J. Med. Chem. 2007, 50, 4087–4095. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.H.; Wu, K.L.; Zhang, X.; Deng, S.Q.; Peng, B. In Silico Screening of Chinese Herbal Medicines With the Potential to Directly Inhibit 2019 Novel Coronavirus. J. Integr. Med. 2020, 18, 152–158. [Google Scholar] [CrossRef]
- Gupta, M.K.; Vemula, S.; Donde, R.; Gouda, G.; Behera, L.; Vadde, R. In-silico Approaches to Detect Inhibitors of the Human Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef] [Green Version]
- Boopathirajan, P.M.K.; Vijayakumar, K. In-Silico Drug Discovery for Covid19 by Targeting Spike Glycoprotein of SARS COV 2 (Wuhan Corona Virus 2019 Outbreak) Against the Docking Analysis with Structure Predicted Human ‘ACE2-FC Region of IGG1’ Fusion Protein As a Protein Based Drug. IJCRT 2020, 8, 1667–1675. [Google Scholar]
- Cava, C.; Bertoli, G.; Castiglioni, I. In Silico Discovery of Candidate Drugs against Covid-19. Viruses 2020, 12, 404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical Predictors of Mortality Due to COVID-19 Based on an Analysis of Data of 150 Patients from Wuhan, China. Intensive Care Med. 2020, 846–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Rosée, P.; Horne, A.; Hines, M.; von Bahr Greenwood, T.; Machowicz, R.; Berliner, N.; Birndt, S.; Gil-Herrera, J.; Girschikofsky, M.; Jordan, M.B.; et al. Recommendations for the Management of Hemophagocytic Lymphohistiocytosis in Adults. Blood 2019, 133, 2465–2477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Ven, A.J.A.M.; Netea, M.G.; van der Meer, J.W.M.; de Mast, Q. Ebola Virus Disease Has Features of Hemophagocytic Lymphohistiocytosis Syndrome. Front. Med. 2015, 2, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huet, T.; Beaussier, H.; Voisin, O.; Jouveshomme, S.; Dauriat, G.; Lazareth, I.; Sacco, E.; Naccache, J.M.; Bézie, Y.; Laplanche, S.; et al. Anakinra for Severe Forms of COVID-19: A Cohort Study. Lancet Rheumatol. 2020. [Google Scholar] [CrossRef]
- King, A.; Vail, A.; O’Leary, C.; Hannan, C.; Brough, D.; Patel, H.; Galea, J.; Ogungbenro, K.; Wright, M.; Pathmanaban, O.; et al. Anakinra in COVID-19: Important Considerations for Clinical Trials. Lancet Rheumatol. 2020, 2, 20–22. [Google Scholar] [CrossRef]
- Calina, D.; Docea, A.O.; Petrakis, D.; Egorov, A.M.; Ishmukhametov, A.A.; Gabibov, A.G.; Shtilman, M.I.; Kostoff, R.; Carvalho, F.; Vinceti, M.; et al. Towards Effective COVID-19 Vaccines: Updates, Perspectives and Challenges (Review). Int. J. Mol. Med. 2020. [Google Scholar] [CrossRef]
- Bloch, E.M.; Shoham, S.; Casadevall, A.; Sachais, B.S.; Shaz, B.; Winters, J.L.; van Buskirk, C.; Grossman, B.J.; Joyner, M.; Henderson, J.P.; et al. Deployment of Convalescent Plasma for the Prevention and Treatment of COVID-19. J. Clin. Investig. 2020, 138745. [Google Scholar] [CrossRef] [Green Version]
- Casadevall, A.; Pirofski, L.A. The Convalescent Sera Option for Containing COVID-19. J. Clin. Investig. 2020, 130, 1545–1548. [Google Scholar] [CrossRef] [Green Version]
- U.S. Food and Drug Administration. Recommendations for Investigational COVID-19 Convalescent Plasma | FDA. Available online: https://www.fda.gov/vaccines-blood-biologics/investigational-new-drug-ind-or-device-exemption-ide-process-cber/recommendations-investigational-covid-19-convalescent-plasma (accessed on 4 May 2020).
- Alattar, R.; Ibrahim, T.; Shaar, S.H.; Abdalla, S.; Shukri, K.; Daghfal, J.N.; Khatib, M.Y.; Aboukamar, M.; Abukhattab, M.; Alsoub, H.A.; et al. Tocilizumab for the Treatment of Severe Coronavirus Disease 2019. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Jalkanen, J.; Hollmén, M.; Jalkanen, S. Interferon Beta-1a for COVID-19: Critical Importance of the Administration Route. Crit. Care 2020, 24, 335. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, H.K.; Mehra, M.R. COVID-19 Illness in Native and Immunosuppressed States: A Clinical-Therapeutic Staging Proposal. J. Heart Lung Transpl. 2020, 39, 405–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients with COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Dastan, F.; Nadji, S.A.; Saffaei, A.; Marjani, M.; Moniri, A.; Jamaati, H.; Hashemian, S.M.; Baghaei, P.; Abedini, A.; Varahram, M.; et al. Subcutaneous Administration of Interferon Beta-1a for COVID-19: A Non-Controlled Prospective Trial. Int. Immunopharmacol. 2020, 85, 106688. [Google Scholar] [CrossRef]
- Hung, I.F.N.; Lung, K.C.; Tso, E.Y.K.; Liu, R.; Chung, T.W.H.; Chu, M.Y.; Ng, Y.Y.; Lo, J.; Chan, J.; Tam, A.R.; et al. Triple Combination of Interferon Beta-1b, Lopinavir-Ritonavir, and Ribavirin in the Treatment of Patients Admitted to Hospital with COVID-19: An Open-Label, Randomised, Phase 2 Trial. Lancet 2020, 395, 1695–1704. [Google Scholar] [CrossRef]
- Tang, N.; Bai, H.; Chen, X.; Gong, J.; Li, D.; Sun, Z. Anticoagulant Treatment Is Associated With Decreased Mortality in Severe Coronavirus Disease 2019 Patients with Coagulopathy. J. Thromb. Haemost. 2020, 18, 1094–1099. [Google Scholar] [CrossRef]
- Connors, J.M.; Levy, J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood 2020. [Google Scholar] [CrossRef]
- Song, J.C.; Wang, G.; Zhang, W.; Zhang, Y.; Li, W.Q.; Zhou, Z.; People’s Liberation Army Professional Committee of Critical Care Medicine, Chinese Society on Thrombosis and Haemostasis. Chinese Expert Consensus on Diagnosis and Treatment of Coagulation Dysfunction in COVID-19. Mil. Med. Res. 2020, 7, 19. [Google Scholar] [CrossRef] [Green Version]













| Clinical Status | Belgium | Italy (Lombardia Protocol) | France | Netherlands | Switzerland | Romania |
|---|---|---|---|---|---|---|
| Mild-to-moderate—no risk group | Symptomatic care (paracetamol) No antiviral treatment | No antiviral treatment | No antiviral treatment | No antiviral treatment | No antiviral treatment | Ambulatory symptomatic care—paracetamol—500 mg/3x/day Hospital treatment—HCQ—400 mg/2x/day—only on day 1 and 200 mg/2x/day—5–6 days Second choice—LPV/r—400/100 mg/2x/day—5–7 days |
| Mild-to-moderate—risk group | HCQ 400 mg at suspicion/diagnosis; 400 mg—12 h later and 200 mg—until day 5—in the absence of HCQ consider CQ base (600 mg/diagnosis; 300 mg—12 h later and 300 mg up to 5 days) or CQ phosphate (1000 mg/diagnosis; 500 mg—12 h later and 300 mg up to 5 days) | LPV/r (400/100 mg/BD) + CQ (500 mg/BD) or HCQ (200 mg/BD)—5–7 days | LPV/r (400/100 mg/BD) (under consideration) Treatment period—depending on the viral excretion | CQ—5 days (under consideration day 1: 600–300 mg; days 2–5: 300 mg) | Not mentioned | HCQ (400 mg/2x/day—only on day 1 and 200 mg/2x/day—4 days) + LPV/r (400/100 mg/2x/day)—10–14 days |
| Severe disease | HCQ—400 mg at suspicion/diagnosis; 400 mg—12 h later and 200 mg—until day 5—in the absence of HCQ consider CQ base (600 mg/diagnosis; 300 mg—12 h later and 300 mg up to 5 days) or CQ phosphate (1000 mg/diagnosis; 500 mg—12 h later and 300 mg up to 5 days) Second option: LPV/r 400/100 mg (=2 tablets of 200/50 mg) BD for 14 days | RDV (200 mg/day—day 1 followed by 100 mg/day days 2–10) + CQ (500 mg/BD) or HCQ (200 mg/BD)—5–20 days (in the absence of RDV, it can be maintained LPV/r+ CQ) | RDV (200 mg/day—day 1 followed by 100 mg/day days 2–10) Treatment period—depending on the viral excretion No second option | CQ—5 days (day 1: 600–300 mg; days 2–5: 300 mg) LPV/r (400/100 mg/BD) as second choice—10–14 days | LPV/r (400/100 mg/BD) (atazanavir/ritonavir as second option) | HCQ (400 mg/2x/days—only on day 1 and 200 mg/2x/day—4–20 days) + RDV (200 mg/day—only on day 1, followed by 100 mg/day for other 9 days) Second choice for RDV is LPV/r, but only until RDV is obtained. ± Tocilizumab (only to patient that present “cytokine storm” and organ dysfunctions)—8 mg/kg body weight (maximum 800 mg)—lent perfusion—1–3 doses at 8 h interval |
| Critical disease | RDV (compassionate use)
| RDV (200 mg/day—day 1 followed by 100 mg/day days 2–10) + CQ (500 mg/BD) or HCQ (200 mg/BD)—5–20 days (in the absence of RDV, it can be maintained LPV/r+ CQ) | RDV (200 mg/day—day 1 followed by 100 mg/day days 2–10) Treatment period—depending on the viral excretion LPV/r—as second option (case by case) | RDV (for 10 days—200 mg/day—day 1 followed by 100 mg/day days 2–10) + CQ (for 5 days—day 1: 600–300 mg; days 2–5: 300 mg) | RDV—10 days (200 mg/day—day 1 followed by 100 mg/day days 2–10) LPV/r (400/100 mg/BD) (+HCQ if <65 years/no comorbidity) as second choice (if RDV is unavailable). Tocilizumab (in the case of MOF and inotropic support) |
| Drug Name | Pharmacological Class | Clinical Phase | EC50 (half maximal effective concentration) | Dose | Mechanism of Action | Adverse Effects |
|---|---|---|---|---|---|---|
| Remdesivir—(RDV) | nucleoside analogue | Severe | 0.77 µM | 200 mg—day 1; 100 mg/day—9 days | inhibitor of the CoVs RNA-dependent RNA polymerase (RdRp) |
|
| Chloroquine (CQ) | 4-aminoquinoline | Mild-to-moderate and severe—depending on the guideline applied | 23.90 µM (24 h) 5.47 µM (48 h) | CQ base (600 mg/diagnosis; 300 mg—12 h later and 300 mg up to 5 days) or CQ phosphate (1000 mg/diagnosis; 500 mg—12 h later and 300 mg up to 5 days) | weak base able to elevate the pH of acidic intracellular organelles, such as endosomes and lysosomes |
|
| Hydroxychloroquine (HCQ) | 4-aminoquinoline | Mild-to-moderate and severe—depending on the guideline applied | 6.14 µM (24 h) 0.72 µM (48 h) | HCQ—400 mg at suspicion/diagnosis; 400 mg—12 h later and 200 mg—until day 5 | weak bases able to elevate the pH of acidic intracellular organelles, such as endosomes and lysosomes | |
| Liponavir/ritonavir (LPv/r) | Protease inhibitor | Mild-to-moderate | - | 400 mg/100 mg/day—14 days | peptidomimetic inhibitor of HIV protease enzyme |
|
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dehelean, C.A.; Lazureanu, V.; Coricovac, D.; Mioc, M.; Oancea, R.; Marcovici, I.; Pinzaru, I.; Soica, C.; Tsatsakis, A.M.; Cretu, O. SARS-CoV-2: Repurposed Drugs and Novel Therapeutic Approaches—Insights into Chemical Structure—Biological Activity and Toxicological Screening. J. Clin. Med. 2020, 9, 2084. https://doi.org/10.3390/jcm9072084
Dehelean CA, Lazureanu V, Coricovac D, Mioc M, Oancea R, Marcovici I, Pinzaru I, Soica C, Tsatsakis AM, Cretu O. SARS-CoV-2: Repurposed Drugs and Novel Therapeutic Approaches—Insights into Chemical Structure—Biological Activity and Toxicological Screening. Journal of Clinical Medicine. 2020; 9(7):2084. https://doi.org/10.3390/jcm9072084
Chicago/Turabian StyleDehelean, Cristina Adriana, Voichita Lazureanu, Dorina Coricovac, Marius Mioc, Roxana Oancea, Iasmina Marcovici, Iulia Pinzaru, Codruta Soica, Aristidis M. Tsatsakis, and Octavian Cretu. 2020. "SARS-CoV-2: Repurposed Drugs and Novel Therapeutic Approaches—Insights into Chemical Structure—Biological Activity and Toxicological Screening" Journal of Clinical Medicine 9, no. 7: 2084. https://doi.org/10.3390/jcm9072084