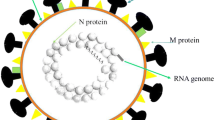
Abstract
Coronavirus disease (COVID-19) is an emerging pandemic that threatens the world since the early days of 2020. Development of vaccines or new drugs against COVID-19 comprises several stages of investigation including efficacy, safety, and approval studies. A shortcut to this delicate pathway is computational-based analysis of FDA-approved drugs against assigned molecular targets of the coronavirus. In this study, we virtually screened a library of FDA-approved drugs prescribed for different therapeutic purposes against versatile COVID-19 specific proteins which are crucial for the virus life cycle. Three antibiotics in our screening polymyxin B, bafilomycin A, and rifampicin show motivating binding stability with more than one target of the virus. Another category of tested drugs is oral antiseptics of mouth rinsing solutions that unexpectedly exhibited significant affinity to the target proteins employed by the virus for attachment and cell internalization. Other OTC drugs widely used and tested in our study are heartburn drugs and they show no significant binding. We tested also some other drugs falling under the scope of investigation regarding interference with a degree of severity of COVID-19 like angiotensin II blockers used as antihypertensive, and our study suggests a therapeutic rather than predisposing effect of these drugs against COVID-19.
Graphical abstract

Similar content being viewed by others
Introduction
Since the early days of 2020, the world is gripped by a COVID-19 pandemic caused by the highly contagious acute respiratory syndrome coronavirus 2 (SARS CoV-2). First, an unknown pneumonia case was detected in Wuhan, province of Hubei in China on December 12, 2019, then a frequent number of cases complained of the related symptoms of dry cough, fever, shortness of breath, and fatigue, that quickly progressed into pneumonia ending up with acute respiratory distress syndrome (ARDS) in 5% of the patients (Chen et al. 2020; Chan et al. 2020; Zhou et al. 2020; Huang et al. 2020). On January 7, the Chinese Centers for Disease Control and Prevention (CDC) announced that a novel coronavirus was identified after whole-genome sequencing of throat swabs and samples obtained from the lower respiratory tract of patients (Zhu et al. 2020; Lu et al. 2020). In February 2020, the World Health Organization (WHO) named it coronavirus disease-19 (COVID-19) officially, then it declared a pandemic on March 12, 2020 (World Health Organization 2020).
By the 3rd of May, 2021, the total number of cases diagnosed with COVID-19 in the world exceeded 153 million, while the total number of recovered cases was more than 131 million, whereas more than 3,200,000 deaths were reported (Coronavirus Update (Live): https://www.worldometers.info/coronavirus - Worldometer, 2021).
In response to this extraordinary pandemic, worldwide research institutes are sprinting to find effective drugs or potential vaccines for COVID-19. There are over 150 vaccines and more than 300 drugs that are under investigation for COVID-19; however, till now, no approved treatments were fully protocolled; on the other hand, mixed point of views regarding efficacy and safety conflict the recently developed vaccines (Pooladanda et al. 2020; Mullard 2020; Sharpe et al. 2020; Thanh Le et al. 2020; Knoll and Wonodi 2020; Polack et al. 2020; Ramasamy et al. 2020; Voysey et al. 2020). Few vaccines have been successfully developed against COVID-19 (Forni and Mantovani 2021), but the major challenge is the production of sufficient doses enough to vaccinate at least 50% of the world population in one hand and the fair distribution among countries on the other hand, aside from the emergence of mutated strains of SARS-CoV-2 virus in some countries like the UK, South Africa, Brazil, and India which may require further investigation to examine the effectiveness of the developed vaccines against the emerged mutants of COVID-19 (Gómez et al. 2021; Hu et al. 2021; Wise 2020; Tegally et al. 2020). All the abovementioned challenges necessitate the design of potential and specific anti-COVID-19 drugs.
Computational aided analysis and virtual screening through molecular docking of FDA-approved drugs rescued long way of research to design novel drugs particularly during periods of emerging pandemics, saved the time required for safety and approval studies, and widened the data base available for screening (Murgueitio et al. 2012). Molecular docking has been employed in structure-based drug design against emerging viruses (Elhefnawi et al. 2012; Plewczynski et al. 2007; Zhou et al. 2008; Raj and Varadwaj 2016).
Based on reported or repurposed anti COVID-19 effects of similar antiparasitic, antimicrobial drugs or drugs that belong to the same medicinal or chemical category like macrolides (Caly et al. 2020; Paccoud et al. 2020; Gautret et al. 2020; Pani et al. 2020; Mahase 2020; Meyerowitz et al. 2020; Ren et al. 2020; Malek et al. 2020), we selected 20 drugs that were not previously assigned by molecular docking against COVID-19. We included in our study some OTC drugs prescribed routinely for heartburn like ranitidine and famotidine, or a common constituent as mucolytics in cough syrups or oral antiseptics in lozenges and mouth washes like ambroxol, bromhexine, chlorhexidine, domiphen, and dequalinium, since many queries are coming up regarding possible protective or medicinal effect of these chemicals against SARS-CoV-2 virus (Carrouel et al. 2020; Freedberg et al. 2020; Janowitz et al. 2020; Sen Gupta et al. 2020). In the same screening, we investigated competitive angiotensin II receptor type 1 (ACE2 T1) antagonists losartan and valsartan that have been widely used for treatment of high blood pressure, since the SARS-CoV2 virus binds to the ACE2 receptors, and more concerns come up regarding the susceptibility of patients using this kind of drugs to the novel coronavirus, SARS-CoV-2 (Annweiler et al. 2020; Gurwitz 2020; Magrone et al. 2020; Wu et al. 2020; Marin 2020).
Methodology
Three-dimensional (3D) structures of all tested compounds (Table 1) were drawn into ChemBioOffice suite, energy minimized to the lowest energy conformer; the crystallized protein structures of the novel coronavirus including COVID-19 main protease (PDB ID:5R7Y) (Douangamath et al. 2020), novel coronavirus spike receptor-binding domain (PDB ID:6LZG) (Wang et al. 2020), prefusion 2019-nCoV spike glycoprotein (PDB ID:6VSB) (Wrapp et al. 2020), and the non structural protein complex NSP16-NSP10 Complex (PDB ID:6W4H) (Rosas-Lemus et al. 2020) were obtained from the protein data bank. The docking process was done using Molecular Operating Environment software (MOE). The compounds were prepared with the standard protocol designated in MOE 2019.01. However, the energy of the docked structures was minimized using MMF94FX forcefield with a gradient RMS of 0.001 kcal/mol, then the protein structure was prepared by using the MOE LigX protocol. The ligands were then docked in the binding site using the Triangle Matcher placement method with 100 runs for each ligand. Refinement was carried out using London dG Forcefield and scored using the GBVI/WSA dG scoring system. Finally, the resulting docking poses were visually inspected, and the pose of the lowest binding free energy value was considered.
Results and discussion
We investigated in silico binding of these collection of drugs against crucial proteins implicated in the life cycle of the novel coronavirus including COVID-19 main protease 5R7Y essential for processing the polyproteins that are translated from the viral RNA, novel coronavirus spike receptor-binding domain 6LZG essential for attaching the virus particles to host cell receptors, prefusion 2019-nCoV spike glycoprotein 6VSB that initiates virus internalization by fusing virus membrane to host cell membrane, and the non structural protein complex NSP16-NSP10 Complex 6W4H from SARS-CoV-2 that plays an important role in virus genome replication and evasion from innate immunity.
Docking of the selected drugs against the COVID-19 main protease enzyme 5R7Y showed the best binding for the cyclic non ribosomal antibiotic polymyxin B with the highest docking score followed by the macrocyclic polyketide antibiotic rifampicin, then the quaternary ammonium compound dequalinium chloride (Fig. 1). Also, the biguanide chlorhexidine and the macrolide antibiotic bafilomycin A1 showed good binding as visualized by the predicted binding modes (Fig. 1), while isoniazid and levamisole showed the least binding score with the target receptor (Table 2). Analysis of modeled protein-ligand architecture of the best fit orientation allowed to recognize the specific amino acids involved in the ligand binding to get this stable complex; the binding simulation of the high score tested compounds uncovered a package of polar amino acids including Thr45, Ser 46, Thr25, Cys44, His41, and Met49 that participate in these interactions and constitute the binding pocket; most of the formed complexes are stabilized via hydrogen bonding (Fig. 1). In line with our findings, polymyxin B, rifampicin, and chlorhexidine have been repurposed as SARS-CoV-2 main proteinase inhibitors in other studies that followed comparable in silico approaches (Maffucci and Contini 2020; Pathak et al. 2020; Sette-De-Souza et al., 2021).
Looking at the docking simulation of the tested drugs against the coronavirus spike receptor-binding domain complexed with ACE2 receptor 6LZG, by analyzing the binding scores for the best fitting poses of each drug (Table 2), we found that polymyxin B again yielded a complex with the highest binding affinity, followed by chlorhexidine, bafilomycin A1, and rifampicin (Fig. 2). Amino acids Gln102, Asn210, Gln98, and Glu208 were repeatedly detected in the binding models of different ligands. Preferential binding of polymyxin B and chlorhexidine was detected in similar studies (Maffucci and Contini 2020; Sette-De-Souza et al., 2021).
Next, we performed further docking simulation against the essential internalization factor Prefusion 2019-nCoV spike glycoprotein 6VSB. According to the docking scores observed for the best fitting orientation (Table 2), the ligands order regarding complex stability almost mimic binding complexes obtained with the spike receptor-binding domain 6LZG exhibiting the following order polymyxin B, rifampicin, bafilomycin A, and chlorhexidine (Fig. 3), which spot the light on possible roles of these drugs to block virus attachment through binding the viral spike proteins and virus internalization. The other drugs showed moderate to mild binding with the mentioned protein. The interacting amino acids that are mostly involved in the ligand binding include Ala706, Asn717, Gln1071, Glu1072, and Asn1074.
Finally, we extended our docking screen of the twenty drugs against the Crystal Structure of NSP16-NSP10 Complex from SARS-CoV-2 6W4H employed by the virus to escape from the immune system. Several drugs showed good binding with the assigned protein; however, the highest docking scores were calculated for polymyxin B followed by chlorhexidine, dequalinium chloride, valsartan, rifampicin, and bafilomycin A1 (Fig. 4); the amino acid interactions that support the predicted binding include Asn6841, Tyr6930, Tyr6845, Gly6871, Cyc6913, Ala6870, and Gly6869 that configure the binding pocket through hydrogen bonding and arene-arene stacking (Fig. 4). While most of the screening approaches designed for COVID-19 commonly test the binding affinity of several candidates against a single COVID-19 substrate, our study utilized a wider approach where the same candidate is tested against more than one substrate; the selected functional proteins play key roles in the life cycle of the virus from the entry and attachment to the cell membrane to virus nucleic acid replication and virus maturation. Our study mapped novel sites of interaction that were not identified by other studies (Maffucci and Contini 2020; Pathak et al. 2020; Sette-De-Souza et al., 2021). Based on our results, we hypothesized that polymyxin B, the most promising candidate together with chlorhexidine in our screening, exhibits different action mechanisms against SARS-CoV-2: (i) acting on viral maturation; (ii) neutralizing spike glycoprotein, preventing the attachment to the ACE-II receptors and virus internalization; and (iii) acting on virus replication. All these mechanisms may decrease pathogenicity in coronaviruses. Bafilomycin A1 appeared among the top 5 candidates that exhibit promising affinity towards all the docked proteins (Table 2); similarly, other studies found the same affinity of bafilomycin A1 against the same COVID-19 targets (Peele et al. 2021);, on the other hand, other in vivo studies attribute the ability of bafilomycin A1 to suppress SARS-CoV-2 replication to its role as an inhibitor of endosomal acidification and blocking endocytic uptake of the virus (Shang et al. 2021). Finally and unexpectedly, the antihypertensive drug valsartan that belongs to the angiotensin II receptor blockers shows a significant affinity to virus proteins that are required for replication and avoidance of innate immunity.
Our results while theoretically reveal unusual affinity of some of these therapeutic compounds against SARS-CoV-2 structural proteins and enzymes, further studied by in vitro and in vivo assays, are essential to examine possible effectiveness.
Conclusions
The COVID-19 pandemic has unified the universal efforts to explore the underlying mechanisms behind virus transmission, virulence, replication, and pathogenesis. Most research pipelines focused on the identification of drug able or vaccine able viral proteins; however, until now, still, no approved drugs were licensed. Current protocols employed for COVID-19 depend on previously designed drugs for closely related viruses or recently repurposed drugs such as chloroquine derivatives, azithromycin, and ivermectin. Following a similar approach, we performed virtual screening of a library of FDA-approved drugs; our study discriminated three promising macrocyclic antibiotics polymyxin B, bafilomycin A, and rifampicin that show promising and consistent in silico binding to more than one protein target of SARS-CoV-2, while other tested antimicrobials that belong to different categories like antituberculosis drugs or antiprotozoal drugs did not show comparable affinity against the same targets. Interestingly, the oral antiseptic chlorhexidine exhibits profound binding affinity to viral proteins especially those required for virus attachments, internalization, and maturation; taken together, our study suggests a protective role of oral antiseptics and mouth rinsing and gargles containing chlorhexidine against COVID-19; on the other hand, OTC drugs for heartburn like ranitidine and famotidine did not show interesting binding scores while being investigated for possible curative effect for COVID-19. Collectively, our findings, while virtual and require further in vitro and in vivo studies for validation, still provide a scientific basis for observational studies and population screening that reports a significant relationship between routine use of some of these tested medications and infection or prognosis of COVID-19.
Data availability
Not applicable
References
Annweiler C, Cao Z, Wu Y, Faucon E, Mouhat S, Kovacic H, Sabatier J-M (2020) Counter-regulatory ‘Renin-Angiotensin’ system-based candidate drugs to treat COVID-19 diseases in SARS-CoV-2-infected patients. Infect Disord Drug Targets 20:407–408. https://doi.org/10.2174/1871526520666200518073329
Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM (2020) The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir Res 178:104787
Carrouel F, Conte MP, Fisher J, Gonçalves LS, Dussart C, Llodra JC, Bourgeois D (2020) COVID-19: a recommendation to examine the effect of mouthrinses with β-cyclodextrin combined with citrox in preventing infection and progression. J Clin Med 9(4). https://doi.org/10.3390/jcm9041126
Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, Xing F, Liu J, Yip CC-Y, Poon RW-S, Tsoi H-W, Lo SK-F, Chan K-H, Poon VK-M, Chan W-M, Ip JD, Cai J-P, Cheng VC-C, Chen H et al (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet (London, England) 395(10223):514–523. https://doi.org/10.1016/S0140-6736(20)30154-9
Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J'a YT, Zhang X, Zhang L (2020) Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (London, England) 395(10223):507–513. https://doi.org/10.1016/S0140-6736(20)30211-7
COVID live update: 153,582,535 cases and 3,218,212 deaths from the coronavirus - Worldometer (2021). https://www.worldometers.info/coronavirus/. Accessed 03 May 2021
Douangamath A, Fearon D, Gehrtz P, Krojer T, Lukacik P, Owen CD, Resnick E, Strain-Damerell C, Aimon A, Ábrányi-Balogh P, Brandão-Neto J, Carbery A, Davison G, Dias A, Downes TD, Dunnett L, Fairhead M, Firth JD, Jones SP et al (2020) Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat Commun 11(1):5047. https://doi.org/10.1038/s41467-020-18709-w
Elhefnawi M, ElGamacy M, Fares M (2012) Multiple virtual screening approaches for finding new hepatitis C virus RNA-dependent RNA polymerase inhibitors: structure-based screens and molecular dynamics for the pursue of new poly pharmacological inhibitors. BMC Bioinformatics 13(Suppl 17):S5. https://doi.org/10.1186/1471-2105-13-S17-S5
Forni G, Mantovani A (2021) COVID-19 vaccines: where we stand and challenges ahead. Cell Death Differ 28(2):626–639. https://doi.org/10.1038/s41418-020-00720-9
Freedberg DE, Conigliaro J, Wang TC, Tracey KJ, Callahan MV, Abrams JA, Sobieszczyk ME, Markowitz DD, Gupta A, O’Donnell MR, Li J, Tuveson DA, Jin Z, Turner WC, Landry DW (2020) Famotidine use is associated with improved clinical outcomes in hospitalized COVID-19 patients: a propensity score matched retrospective cohort study. Gastroenterology 159:1129–1131.e3. https://doi.org/10.1053/j.gastro.2020.05.053
Gautret P, Lagier J-C, Parola P, van Hoang T, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira VE, Dupont HT, Honoré S, Colson P, Chabrière E, La Scola B, Rolain J-M, Brouqui P, Raoult D (2020) Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 56(1):105949. https://doi.org/10.1016/j.ijantimicag.2020.105949
Gómez CE, Perdiguero B, Esteban M (2021) Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccines 9(3):243. https://doi.org/10.3390/vaccines9030243
Gurwitz D (2020) Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev Res 81:537–540. https://doi.org/10.1002/ddr.21656
Hu J, Peng P, Wang K, Fang L, Luo F-y, A-s J, B-z L, Tang N, A-l H (2021) Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell Mol Immunol 18(4):1061–1063. https://doi.org/10.1038/s41423-021-00648-1
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506. https://doi.org/10.1016/S0140-6736(20)30183-5
Janowitz T, Gablenz E, Pattinson D, Wang TC, Conigliaro J, Tracey K, Tuveson D (2020) Famotidine use and quantitative symptom tracking for COVID-19 in non-hospitalised patients: a case series. Gut 69:1592–1597. https://doi.org/10.1136/gutjnl-2020-321852
Knoll MD, Wonodi C (2020) Oxford–AstraZeneca COVID-19 vaccine efficacy. Lancet 397:72–74. https://doi.org/10.1016/S0140-6736(20)32623-4
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (London, England) 395(10224):565–574. https://doi.org/10.1016/S0140-6736(20)30251-8
Maffucci I, Contini A (2020) In silico drug repurposing for SARS-CoV-2 main proteinase and spike proteins. J Proteome Res 19(11):4637–4648. https://doi.org/10.1021/acs.jproteome.0c00383
Magrone T, Magrone M, Jirillo E (2020) Focus on receptors for coronaviruses with special reference to angiotensin-converting enzyme 2 as a potential drug target - a pserspective. Endocr Metab Immune Disord Drug Targets 20:807–811. https://doi.org/10.2174/1871530320666200427112902
Mahase E (2020) Hydroxychloroquine for covid-19: the end of the line? BMJ (Clin Res Ed) 369:m2378. https://doi.org/10.1136/bmj.m2378
Malek AE, Granwehr BP, Kontoyiannis DP (2020) Doxycycline as a potential partner of COVID-19 therapies. IDCases 21:e00864. https://doi.org/10.1016/j.idcr.2020.e00864
Marin GH (2020) Facts and reflections on COVID-19 and anti-hypertensives drugs. Drug Discov Ther 14(2):105–106. https://doi.org/10.5582/ddt.2020.01017
Meyerowitz EA, Vannier AGL, Friesen MGN, Schoenfeld S, Gelfand JA, Callahan MV, Kim AY, Reeves PM, Poznansky MC (2020) Rethinking the role of hydroxychloroquine in the treatment of COVID-19. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 34(5):6027–6037
Mullard A (2020) Flooded by the torrent: the COVID-19 drug pipeline. Lancet (London, England) 395(10232):1245–1246. https://doi.org/10.1016/S0140-6736(20)30894-1
Murgueitio MS, Bermudez M, Mortier J, Wolber G (2012) In silico virtual screening approaches for anti-viral drug discovery. Drug discovery today. Technologies 9(3):e219–e225. https://doi.org/10.1016/j.ddtec.2012.07.009
Paccoud O, Tubach F, Baptiste A, Bleibtreu A, Hajage D, Monsel G, Tebano G, Boutolleau D, Klement E, Godefroy N, Palich R, Itani O, Fayssal A, Valantin M-A, Tubiana R, Burrel S, Calvez V, Caumes E, Marcelin A-G, Pourcher V (2020) Compassionate use of hydroxychloroquine in clinical practice for patients with mild to severe Covid-19 in a French university hospital. Clin Infect Dis 18:ciaa791. https://doi.org/10.1093/cid/ciaa791
Pani A, Lauriola M, Romandini A, Scaglione F (2020) Macrolides and viral infections: focus on azithromycin in COVID-19 pathology. Int J Antimicrob Agents 56:106053. https://doi.org/10.1016/j.ijantimicag.2020.106053
Pathak Y, Mishra A, Tripathi V (2020) Rifampicin may be repurposed for COVID-19 treatment: insights from an in-silico study. PREPRINT (Version 1) available at Research Square. https://doi.org/10.21203/rs.3.rs-22546/v1
Peele KA, Kumar V, Parate S, Srirama K, Lee KW, Venkateswarulu TC (2021) In silico drug repurposing using FDA approved drugs against membrane protein of SARS-CoV-2. J Pharm Sci 110:2346–2354. https://doi.org/10.1016/j.xphs.2021.03.004
Plewczynski D, Hoffmann M, von Grotthuss M, Ginalski K, Rychewski L (2007) In silico prediction of SARS protease inhibitors by virtual high throughput screening. Chem Biol Drug Des 69(4):269–279. https://doi.org/10.1111/j.1747-0285.2007.00475.x
Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL et al (2020) Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 383:2603–2615. https://doi.org/10.1056/NEJMoa2034577
Pooladanda V, Thatikonda S, Godugu C (2020) The current understanding and potential therapeutic options to combat COVID-19. Life Sci 254:117765. https://doi.org/10.1016/j.lfs.2020.117765
Raj U, Varadwaj PK (2016) Flavonoids as multi-target inhibitors for proteins associated with Ebola virus: in silico discovery using virtual screening and molecular docking studies. Interdiscip Sci Comput Life Sci 8(2):132–141. https://doi.org/10.1007/s12539-015-0109-8
Ramasamy MN, Minassian AM, Ewer KJ, Flaxman AL, Folegatti PM, Owens DR, Voysey M, Aley PK, Angus B, Babbage G, Belij-Rammerstorfer S, Berry L, Bibi S, Bittaye M, Cathie K, Chappell H, Charlton S, Cicconi P, Clutterbuck EA et al (2020) Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet, London, England. https://doi.org/10.1016/S0140-6736(20)32466-1
Ren L, Xu W, Overton JL, Yu S, Chiamvimonvat N, Thai PN (2020) Assessment of hydroxychloroquine and chloroquine safety profiles: a systematic review and meta-analysis. medRxiv : the preprint server for health sciences. doi: https://doi.org/10.1101/2020.05.02.20088872
Rosas-Lemus M, Minasov G, Shuvalova L, Inniss NL, Kiryukhina O, Brunzelle J, Satchell KJF (2020) High-resolution structures of the SARS-CoV-2 2′- O -methyltransferase reveal strategies for structure-based inhibitor design. Sci Signal 13(651):eabe1202. https://doi.org/10.1126/scisignal.abe1202
Sen Gupta PS, Biswal S, Singha D, Rana MK (2020) Binding insight of clinically oriented drug famotidine with the identified potential target of SARS-CoV-2. J Biomol Struct Dyn 39(14):5327–5333. https://doi.org/10.1080/07391102.2020.1784795
Sette-DE-Souza PH, Costa MJF, Amaral-Machado L, Araújo FADC, Almeida Filho AT, Lima LRA (2021) Dental workers in front-line of COVID-19: an in silico evaluation targeting their prevention. J Appl Oral Sci 26;29:e20200678. https://doi.org/10.1590/1678-7757-2020-0678
Shang C, Zhuang X, Zhang H, Li Y, Zhu Y, Lu J, Ge C, Cong J, Li T, Tian M, Jin N, Li X (2021) Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice. Virol J 18(1):46. https://doi.org/10.1186/s12985-021-01515-1
Sharpe HR, Gilbride C, Allen E, Belij-Rammerstorfer S, Bissett C, Ewer K, Lambe T (2020) The early landscape of COVID-19 vaccine development in the UK and rest of the world. Immunology 160:223–232. https://doi.org/10.1111/imm.13222
Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, Gottberg A von, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao M, Korsman S, Davies M-A, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Pond SLK, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, Oliveira T de (2020) Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. https://doi.org/10.1101/2020.12.21.20248640
Thanh Le T, Andreadakis Z, Kumar A, Gómez Román R, Tollefsen S, Saville M, Mayhew S (2020) The COVID-19 vaccine development landscape. Nat Rev Drug Discov 19(5):305–306. https://doi.org/10.1038/d41573-020-00073-5
Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Collins AM, Colin-Jones R, Cutland CL, Darton TC, Dheda K, Duncan CJA et al (2020) Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet, London, England. https://doi.org/10.1016/S0140-6736(20)32661-1
Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen K-Y, Wang Q, Zhou H, Yan J, Qi J (2020) Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181(4):894–904.e9. https://doi.org/10.1016/j.cell.2020.03.045
Wise J (2020) Covid-19: new coronavirus variant is identified in UK. BMJ (Clinical research ed.) 371:m4857. https://doi.org/10.1136/bmj.m4857
World Health Organization (2020) Novel coronavirus – China. https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/. Accessed 11 Jun 2020
Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, NY) 367(6483):1260–1263. https://doi.org/10.1126/science.abb2507
Wu C, Ye D, Mullick AE, Li Z, Danser AHJ, Daugherty A, Lu HS (2020) Effects of renin-angiotensin inhibition on ACE2 and TMPRSS2 expression: insights into COVID-19. bioRxiv : the preprint server for biology. doi: https://doi.org/10.1101/2020.06.08.137331
Zhou Z, Khaliq M, Suk J-E, Patkar C, Li L, Kuhn RJ, Post CB (2008) Antiviral compounds discovered by virtual screening of small-molecule libraries against dengue virus E protein. ACS Chem Biol 3(12):765–775. https://doi.org/10.1021/cb800176t
Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. https://doi.org/10.1038/s41586-020-2012-7
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382(8):727–733. https://doi.org/10.1056/NEJMoa2001017
Acknowledgements
The authors would like to extend their sincere appreciation to the Researchers Supporting Project number (RSP-2021/96), King Saud University, Riyadh, Saudi Arabia. GY is grateful to Alexander Von Humboldt (AVH) foundation for post-doc fellowship.
Author information
Authors and Affiliations
Contributions
Conceptualization: Galal Yahya and Ahmed M. El-Baz; methodology: Mohammad Elmorsy; Writing original draft preparation: Ahmed M. El-Baz, Mohammad Elmorsy, Nashwa Mohamed, and Rafa Almeer; Writing, review, and editing: Mohamed Abdel-Daim and Galal Yahya
Corresponding authors
Ethics declarations
Ethical approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Philippe Garrigues
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Elmorsy, .A., El-Baz, A.M., Mohamed, N.H. et al. In silico screening of potent inhibitors against COVID-19 key targets from a library of FDA-approved drugs. Environ Sci Pollut Res 29, 12336–12346 (2022). https://doi.org/10.1007/s11356-021-16427-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-021-16427-4