SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antibodies and Reagents
2.2. Cell Culture, Infection, and Treatments
2.3. RNA Interference
- 5′-GUUCUUCGUGAGUUCCUCdTdT-3′,
- 3′-dTdTCAAGAAGCACUCAAGGAG-5′
2.4. RNA Extraction and Reverse Transcription
2.5. Droplet Digital PCR (ddPCR)
2.6. Western Blotting
3. Results
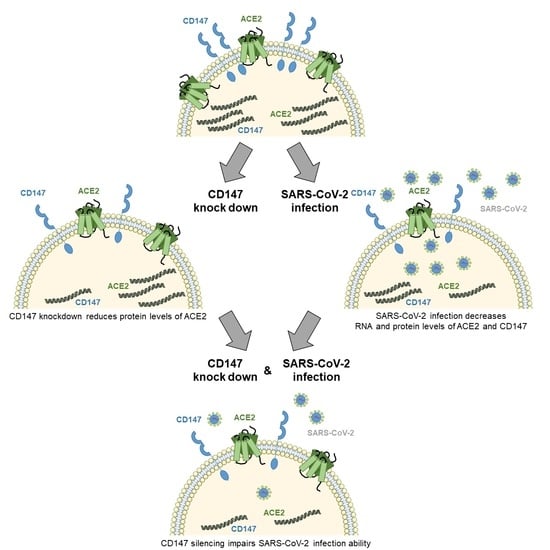
3.1. CD147 Plays a Different Role in SARS-CoV and SARS-CoV-2 Entry
3.2. CD147 Silencing Reduces ACE2 Levels and SARS-CoV-2 Infection
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of Therapeutic Targets for SARS-CoV-2 and Discovery of Potential Drugs by Computational Methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural Basis for the Recognition of the SARS-CoV-2 by Full-Length Human ACE2. Science 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 Facilitates SARS-CoV-2 Cell Entry and Infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 Is a Host Factor for SARS-CoV-2 Infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.-Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; et al. CD147-Spike Protein Is a Novel Route for SARS-CoV-2 Infection to Host Cells. Signal Transduct. Target. Ther. 2020, 5, 1–10. [Google Scholar] [CrossRef]
- Xiong, L.; Edwards, C.K.; Zhou, L. The Biological Function and Clinical Utilization of CD147 in Human Diseases: A Review of the Current Scientific Literature. Int. J. Mol. Sci. 2014, 15, 17411–17441. [Google Scholar] [CrossRef] [Green Version]
- Yurchenko, V.; Zybarth, G.; O’Connor, M.; Dai, W.W.; Franchin, G.; Hao, T.; Guo, H.; Hung, H.-C.; Toole, B.; Gallay, P.; et al. Active Site Residues of Cyclophilin A Are Crucial for Its Signaling Activity via CD147. J. Biol. Chem. 2002, 277, 22959–22965. [Google Scholar] [CrossRef] [Green Version]
- Pushkarsky, T.; Zybarth, G.; Dubrovsky, L.; Yurchenko, V.; Tang, H.; Guo, H.; Toole, B.; Sherry, B.; Bukrinsky, M. CD147 Facilitates HIV-1 Infection by Interacting with Virus-Associated Cyclophilin A. Proc. Natl. Acad. Sci. USA 2001, 98, 6360–6365. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Mi, L.; Xu, J.; Yu, J.; Wang, X.; Jiang, J.; Xing, J.; Shang, P.; Qian, A.; Li, Y.; et al. Function of HAb18G/CD147 in Invasion of Host Cells by Severe Acute Respiratory Syndrome Coronavirus. J. Infect. Dis. 2005, 191, 755–760. [Google Scholar] [CrossRef] [Green Version]
- Shilts, J.; Crozier, T.W.M.; Greenwood, E.J.D.; Lehner, P.J.; Wright, G.J. No Evidence for Basigin/CD147 as a Direct SARS-CoV-2 Spike Binding Receptor. Sci. Rep. 2021, 11, 413. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Chen, Y.; Gou, X.; Ke, X.; Cui, H.; Chen, Z. Human Tumor Cells Induce Angiogenesis through Positive Feedback between CD147 and Insulin-Like Growth Factor-I. PLoS ONE 2012, 7, e40965. [Google Scholar] [CrossRef] [PubMed]
- Fenizia, C.; Biasin, M.; Cetin, I.; Vergani, P.; Mileto, D.; Spinillo, A.; Gismondo, M.R.; Perotti, F.; Callegari, C.; Mancon, A.; et al. Analysis of SARS-CoV-2 Vertical Transmission during Pregnancy. Nat. Commun. 2020, 11, 5128. [Google Scholar] [CrossRef]
- Dittmar, M.; Lee, J.S.; Whig, K.; Segrist, E.; Li, M.; Kamalia, B.; Castellana, L.; Ayyanathan, K.; Cardenas-Diaz, F.L.; Morrisey, E.E.; et al. Drug Repurposing Screens Reveal Cell-Type-Specific Entry Pathways and FDA-Approved Drugs Active against SARS-Cov-2. Cell Rep. 2021, 35, 108959. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, C.; Staffler, G.; Hüttinger, R.; Hilgert, I.; Prager, E.; Černý, J.; Steinlein, P.; Majdic, O.; Hořejší, V.; Stockinger, H. T Cell Activation-Associated Epitopes of CD147 in Regulation of the T Cell Response, and Their Definition by Antibody Affinity and Antigen Density. Int. Immunol. 1999, 11, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A Crucial Role of Angiotensin Converting Enzyme 2 (ACE2) in SARS Coronavirus–Induced Lung Injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Herzog, P.; Pfefferle, S.; Steffen, I.; Muench, M.O.; Simmons, G.; Hofmann, H.; Kuri, T.; Weber, F.; et al. Differential Downregulation of ACE2 by the Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus and Human Coronavirus NL63. J. Virol. 2010, 84, 1198–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Zhou, Y.; Zhang, M.; Wang, H.; Zhao, Q.; Liu, J. Updated Approaches against SARS-CoV-2. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahn, J.N.; Kaushik, D.K.; Yong, V.W. The Role of EMMPRIN in T Cell Biology and Immunological Diseases. J. Leukoc. Biol. 2015, 98, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, V.; Constant, S.; Eisenmesser, E.; Bukrinsky, M. Cyclophilin–CD147 Interactions: A New Target for Anti-inflammatory Therapeutics. Clin. Exp. Immunol. 2010, 160, 305–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Y.-X.; Wang, S.-J.; Fan, J.-H.; Sun, S.-J.; Li, X.; Padhiar, A.A.; Zhang, J.-N. CD147 Stimulates Hepatoma Cells Escaping from Immune Surveillance of T Cells by Interaction with Cyclophilin A. Biomed. Pharm. 2016, 80, 289–297. [Google Scholar] [CrossRef]
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e19. [Google Scholar] [CrossRef]
- Kaushik, D.K.; Bhattacharya, A.; Mirzaei, R.; Rawji, K.S.; Ahn, Y.; Rho, J.M.; Yong, V.W. Enhanced Glycolytic Metabolism Supports Transmigration of Brain-Infiltrating Macrophages in Multiple Sclerosis. J. Clin. Investig. 2019, 129, 3277–3292. [Google Scholar] [CrossRef] [PubMed]
- Philp, N.J.; Ochrietor, J.D.; Rudoy, C.; Muramatsu, T.; Linser, P.J. Loss of MCT1, MCT3, and MCT4 Expression in the Retinal Pigment Epithelium and Neural Retina of the 5A11/Basigin-Null Mouse. Investig. Ophthalmol. Vis. Sci. 2003, 44, 1305–1311. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Wan, C.; Wang, Z.-D.; Gao, Y.; Li, Y.-C.; Tang, F.; Zhu, H.-Y.; Yi, L.-X.; Zhang, C. Expression of CD147 and Cyclophilin A in Kidneys of Patients with COVID-19. Clin. J. Am. Soc. Nephrol. 2020. [Google Scholar] [CrossRef]
- Sims, A.C.; Baric, R.S.; Yount, B.; Burkett, S.E.; Collins, P.L.; Pickles, R.J. Severe Acute Respiratory Syndrome Coronavirus Infection of Human Ciliated Airway Epithelia: Role of Ciliated Cells in Viral Spread in the Conducting Airways of the Lungs. J. Virol. 2005, 79, 15511–15524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deora, A.A.; Gravotta, D.; Kreitzer, G.; Hu, J.; Bok, D.; Rodriguez-Boulan, E. The Basolateral Targeting Signal of CD147 (EMMPRIN) Consists of a Single Leucine and Is Not Recognized by Retinal Pigment Epithelium. Mol. Biol. Cell 2004, 15, 4148–4165. [Google Scholar] [CrossRef] [PubMed]
- Radzikowska, U.; Ding, M.; Tan, G.; Zhakparov, D.; Peng, Y.; Wawrzyniak, P.; Wang, M.; Li, S.; Morita, H.; Altunbulakli, C.; et al. Distribution of ACE2, CD147, CD26, and Other SARS-CoV-2 Associated Molecules in Tissues and Immune Cells in Health and in Asthma, COPD, Obesity, Hypertension, and COVID-19 Risk Factors. Allergy 2020, 75, 2829–2845. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, D.K.; Hahn, J.N.; Yong, V.W. EMMPRIN, an Upstream Regulator of MMPs, in CNS Biology. Matrix Biol. 2015, 44–46, 138–146. [Google Scholar] [CrossRef]
- Von Ungern-Sternberg, S.N.I.; Zernecke, A.; Seizer, P. Extracellular Matrix Metalloproteinase Inducer EMMPRIN (CD147) in Cardiovascular Disease. Int. J. Mol. Sci. 2018, 19, 507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosugi, T.; Maeda, K.; Sato, W.; Maruyama, S.; Kadomatsu, K. CD147 (EMMPRIN/Basigin) in Kidney Diseases: From an Inflammation and Immune System Viewpoint. Nephrol. Dial. Transplant. 2015, 30, 1097–1103. [Google Scholar] [CrossRef] [Green Version]
- Qu, X.; Wang, C.; Zhang, J.; Qie, G.; Zhou, J. The Roles of CD147 and/or Cyclophilin A in Kidney Diseases. Mediat. Inflamm. 2014, 2014, e728673. [Google Scholar] [CrossRef]
- Satoh, K.; Satoh, T.; Kikuchi, N.; Omura, J.; Kurosawa, R.; Suzuki, K.; Sugimura, K.; Aoki, T.; Nochioka, K.; Tatebe, S.; et al. Basigin Mediates Pulmonary Hypertension by Promoting Inflammation and Vascular Smooth Muscle Cell Proliferation. Circ. Res. 2014, 115, 738–750. [Google Scholar] [CrossRef] [Green Version]
- Bian, H.; Zheng, Z.-H.; Wei, D.; Zhang, Z.; Kang, W.-Z.; Hao, C.-Q.; Dong, K.; Kang, W.; Xia, J.-L.; Miao, J.-L.; et al. Meplazumab Treats COVID-19 Pneumonia: An Open-Labelled, Concurrent Controlled Add-on Clinical Trial. MedRxiv 2020. [Google Scholar] [CrossRef]
- Muramatsu, T. Basigin (CD147), a Multifunctional Transmembrane Glycoprotein with Various Binding Partners. J. Biochem. 2016, 159, 481–490. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 Variants to Neutralization by Monoclonal and Serum-Derived Polyclonal Antibodies. Nat. Med. 2021, 1–10. [Google Scholar] [CrossRef]
- Xie, X.; Liu, Y.; Liu, J.; Zhang, X.; Zou, J.; Fontes-Garfias, C.R.; Xia, H.; Swanson, K.A.; Cutler, M.; Cooper, D.; et al. Neutralization of SARS-CoV-2 Spike 69/70 Deletion, E484K and N501Y Variants by BNT162b2 Vaccine-Elicited Sera. Nat. Med. 2021, 620–621. [Google Scholar] [CrossRef] [PubMed]
- Muik, A.; Wallisch, A.-K.; Sänger, B.; Swanson, K.A.; Mühl, J.; Chen, W.; Cai, H.; Maurus, D.; Sarkar, R.; Türeci, Ö.; et al. Neutralization of SARS-CoV-2 Lineage B.1.1.7 Pseudovirus by BNT162b2 Vaccine–Elicited Human Sera. Science 2021, 371, 1152–1153. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E.; et al. Escape from Neutralizing Antibodies by SARS-CoV-2 Spike Protein Variants. Elife 2020, 9, e61312. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Peng, P.; Wang, K.; Liu, B.; Fang, L.; Luo, F.; Jin, A.; Tang, N.; Huang, A. Emerging SARS-CoV-2 Variants Reduce Neutralization Sensitivity to Convalescent Sera and Monoclonal Antibodies. Cell. Mol. Immunol. 2021, 18, 1061–1063. [Google Scholar] [CrossRef] [PubMed]



| Antibody | Company | Catalog | Dilution | Conditions |
|---|---|---|---|---|
| Spike | Genetex | 632,604 | 1:1000 | Overnight, 4 °C |
| ACE2 | Abcam | 15,348 | 1:1000 | Overnight, 4 °C |
| CD147 | Santa Cruz | 53,693 | 1:500 | Overnight, 4 °C |
| CyPA | Abcam | 58,144 | 1:500 | Overnight, 4 °C |
| GAPDH | Abcam | 128,915 | 1:40,000 | Overnight, 4 °C |
| Anti-mouse HRP | Cell Signaling Technologies | 7076 | 1:5000 | 1 h, r.t. |
| Anti-rabbit HRP | Cell Signaling Technologies | 7074 | 1:5000 | 1 h, r.t. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fenizia, C.; Galbiati, S.; Vanetti, C.; Vago, R.; Clerici, M.; Tacchetti, C.; Daniele, T. SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2. Cells 2021, 10, 1434. https://doi.org/10.3390/cells10061434
Fenizia C, Galbiati S, Vanetti C, Vago R, Clerici M, Tacchetti C, Daniele T. SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2. Cells. 2021; 10(6):1434. https://doi.org/10.3390/cells10061434
Chicago/Turabian StyleFenizia, Claudio, Silvia Galbiati, Claudia Vanetti, Riccardo Vago, Mario Clerici, Carlo Tacchetti, and Tiziana Daniele. 2021. "SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2" Cells 10, no. 6: 1434. https://doi.org/10.3390/cells10061434