Abstract
We predict the evolution of the COVID-19 pandemic in several countries using a logistic model. The model uses a regression analysis based on the least-squares fitting. In particular, the growth rate of the infection has been fitted as an exponential decay, as compared to a linear decay, reported previously in logistic models. The model has been validated with the data of China and South Korea, where the pandemic is nearing to its end. The data of Italy, Germany, Spain, and Sweden show that the peak of the infection has been reached, i.e. a time when the new infections will start to decrease as compared to the previous day. The model predicts the approximate number of total infections at the end of the outbreak. The possible peak date and the total number of infections for different countries are predicted using the data available. The total number of infections in the USA is estimated to be around 4 million. The model prediction of Brazil shows that the peak will reach on 5 July 2020 and total infections will be 3.2 million. The reported data of India show a large initial scatter in the growth rate. The total number of infections in India is estimated to be around 2.4 million by the model and the predicted peak date is 3 August 2020. The predictions of India are discussed in the context of restricted movement of population, i.e. lock-down imposed by the government.
Similar content being viewed by others
Introduction
COVID-19 pandemic, caused by the SARS-CoV-2 virus, has affected most of the countries around the world and has presented several challenges to humanity and science. A simple predictive model in this context can help to devise or modify policies by the governments while designing the mitigation measures and to judiciously use the healthcare infrastructure. Several factors influence the evolution of COVID-19, such as movement restriction of people by the government (lock-down), outdoor weather, and change in virulence of virus with time. In a recent study, Bhardwaj and Agrawal (2020) showed that the growth rate is weakly correlated to the outdoor weather during the evolution of the pandemic since the latter influences the drying time of respiratory droplets on a surface. To understand the effect of these factors on the evolution of an infectious disease, a predictive model applied to different countries could help to gain understanding of the evolution since these factors may vary from one country to another.
The epidemiological models are classified as exponential, logistic, and susceptible–infectious–recovered (SIR) models (Renshaw 1993; Ranjan 2020). Previous studies have proposed models in these categories to predict the outcome and evolution of COVID-19 in different countries. The exponential models are useful for predicting the initial growth of an infectious disease (Ranjan 2020). For instance, Verma et al. (2020) used a variant of the exponential model, and used exponential and power-law functions to fit the rate of change of COVID-19 infections in different countries and argued that transition from an exponential regime to a power-law regime is a signature of the flattening of the curve. The logistic models have been used to predict the decay of the infections but such a model may fail in initial stages (Ranjan 2020). Vattay (2020) employed a logistic model for predicting the death rate in Italy and assumed that the growth rate of the infection is linear. Similarly, Batista (2020) employed a logistic growth regression model to predict the total infections and peak time of the pandemic. This study modeled the growth rate as linear decay. It was shown in Quadrat (2020) recently that a SIR model does not capture the data of France and a logistic model is a better candidate to predict the pandemic evolution, specifically when the movement of the population, i.e. a lock-down, is imposed. Ranjan (2020) proposed an exponential and classic SIR model based on the available data and predicted that basic reproduction number (\(R_0\)) for India is in the range of 1.4–3.9. Based on the SIR model, this study predicted that the peak date of infection will occur at the end of May in India if community transmission does not occur.
While several predictive logistic models have been reported previously (e.g. Vattay 2020; Batista 2020; Quadrat 2020), the present modeling effort is to develop a data-driven, extrapolation model that could render more accurate predictions. A previous logistic model used a linear fit for the growth rate of the infection and could not predict the data accurately (Vattay 2020). In the present work, we extend the model in Vattay (2020) and use an exponential function to fit the growth rate of the infection. The present results show that a logistic model, with growth rate modeled as an exponential decay, can confirm and predict the evolution of COVID-19 in several countries with reasonable fidelity.
Methods
The following ordinary differential equation governs the evolution of a disease in given human population (Vattay 2020):
where N is number of people infected at a given time t and \({\lambda }\) is the growth rate of the infections. The model uses reported field data of the infections of a population over a specific time period [\(t_0\), \(t_1\), \(t_2\), .......,\(t_P\)], say [\(N_0\), \(N_1\), \(N_2\), .......,\(N_p\)], where subscripts 0 and p refer to the 0th and present day of the outbreak. The growth rate can be numerically approximated from Eq. 1(Vattay 2020),
Considering the unit of time as day and since the data is available for each day, we use \(\Delta t\) = 1 in the above equation. We estimate \({\lambda }_{t_{i}}\) from the data of different countries given in public domain (https://coronavirus.jhu.edu, https://www.worldometers.info/coronavirus and https://www.covid19india.org), and use a regression analysis based least-squares fitting to the data of \({\lambda }_{t_{i}}\). The fitting results in the present work show that the exponential fit works well with the given data, as compared to a linear fit. The initial scatter in the growth rate was ignored while fitting the growth rate with an exponential function. The start day for the fitting of the curve corresponds to around 1% of the total infections on the present day.
Using the fit obtained, we extrapolate number of infections which will occur in future for \(t > t_p\) using the fitted value of \({\lambda }_{t_{i}}\). The initial value of this extrapolation is taken as \(N_P\). This allows us to predict the time of the peak of infections, i.e. after this time, the daily infections will start to reduce. The total number of infections is also predicted, with the same extrapolation. The peak time is obtained by plotting \(N_{t_{i}} - N_{t_{i-1}}\) against time and the end of the outbreak is considered when \({\lambda }_{t_{i}} \le 0.001-0.01\), i.e. the rise in the total number of infections falls below 0.1–1%. The total infections are estimated at the end of the outbreak.
Model Validation
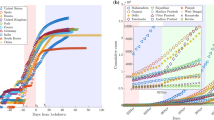
The model has been validated with the data of China and South Korea (https://coronavirus.jhu.edu). These two countries have been selected since the outbreak of the pandemic is nearing to its end. Figures 1 and 2 show that the time-history of the growth rate of the two countries and fitted exponential decays are in good agreement.
Validation of the model for South Korea. Rest of the caption is same as of Fig. 1
Results and Discussion
First, we plot the results of the three countries—-Italy (Fig. 3), Germany (Fig. 4), Spain (Fig. 5) and Sweden (Fig. 6), where the pandemic has reached its peak in March or April 2020 and the growth has slowed down. The data used in the model are from 22 January 2020 to 10 April 2020. Time = 0 corresponds to 22 January 2020 in all figures. The peak date of the infection obtained from the data (https://www.worldometers.info/coronavirus) and the predicted number of infections have been listed for these countries in Table 1. The predicted number of infections is on the same order in Italy, Germany, and Spain. The number of infections in Sweden will be one order of magnitude lesser than Italy, Germany, and Spain, despite no lock-down in the former.
Data and model forecast for Italy. Day 0 is 22 January 2020 and data until 10 April 2020 is analysed. Time-history of the growth rate of the infections per day (top row), cumulative number of infections (middle row) and daily infections (bottom row) are plotted. The data are shown by symbols while the solid lines are predicted from the model. Simulation is stopped if \({\lambda }_{t_{i}} \le 0.01\) or the change in total infections per day is lesser than 1%
Data and model forecast for Germany. Rest of the caption is same as of Fig. 3
Data and model forecast for Spain. Rest of the caption is same as of Fig. 3
Data and model forecast for Sweden. Rest of the caption is same as of Fig. 3
Second, we plot the results of the USA where the peak of the infections has reached in April 2020. The data used in the model are from 22 January 2020 to 10 April 2020. Figure 7 plots the data of the USA and the peak of the pandemic is predicted to be around 03 May 2020. The data available (https://www.worldometers.info/coronavirus) on 1 June 2020 shows that the peak date was 24 April 2020. Therefore, the model predicts the peak date of the USA with reasonable fidelity (with an error of 9 days). The total number of predicted infections by the model in the USA is around 4.2 million (Table 1).
Data and model forecast for USA. Rest of the caption is same as of Fig. 3
Third, we plot the results of countries, Brazil and India, that are expecting the peak of the infections after April 2020. The data used in the model for these two countries are from 22 January 2020 to 31 May 2020. Figure 8 plots the data of Brazil and we note that the predicted peak of the pandemic is on 5 July 2020, with a total number of infections around 3.2 million (Table 2).
Data and model forecast for Brazil. Day 0 is 22 Jan 2020, and data until 31 May 2020 are analyzed. Time-history of the growth rate of the infections per day (top row), cumulative number of infections (middle row) and daily infections (bottom row) are plotted. The data are shown by symbols while the solid lines are predicted from the model. Simulation is stopped if \({\lambda }_{t_{i}} \le 0.001\) or the change in total infections per day is lesser than 0.1%
Finally, the data for India are plotted in Fig. 9. The data of growth rate in Fig. 9 (first row) shows reasonable scatter until around Day 75 and after that, we observe that the growth rate decays exponentially. The model prediction shows that the peak will occur on 3 August 2020 (Table 2), with total infections of around 2.4 × 106. The plot of daily infections against time shows that the curve is a bit broader compared to that of other countries. A complete lock-down, imposed at the starting of the outbreak since 22 March 2020, maybe a reason for the slower outbreak. The initial scatter in the data could be attributed to cluster transmission initially and when community transmission was established, the growth rate decay became smoother after around Day 75. Therefore, a broader peak and slow decay of the growth rate are possible signatures of the effect of a strict lock-down imposed by the government.
Data and model forecast for India. Rest of the caption is the same as of Fig. 8
The model presented here has certain limitations. It assumes a constant human population with uniform mixing of the people and cannot predict the total number of fatalities or recoveries. The recovery from the infections is not considered, typically considered in classical SIR models. The recovery could reduce the active number of infections and thereby the growth rate. The model does not directly account for the mitigation measures, such as movement restriction of the population, i.e. lock-down, taken by the respective governments. However, there is an indirect effect of the lock-down on the growth rate and it is accounted for in the model by assuming it as an exponential decay. The model predictions are based on the reported data, which serves as an essential input to the model. In general, the predictions improve as the time progresses since more data becomes available (essentially, the model is data-driven, like a weather prediction model).
Conclusions
A logistic model to predict the evolution of COVID-19 pandemic is presented. This predictive mathematical model uses a regression analysis based on least-squares fitting. Specifically, the growth rate of the infection is fitted as an exponential decay, as compared to a linear decay. The model validation is carried out with the data of China and South Korea, where the pandemic has ended. We predict the date of peak infections, i.e. a date when the new infections will start to decrease as compared to the previous day and the total number of infections at the end of the outbreak. Data from several countries, namely Italy, Germany, Spain, and Sweden, USA, Brazil, and India are plotted for making predictions. All tested countries show a large initial scatter in the growth rate except Italy. The data of Italy, Germany, Spain, and Sweden show that the peak of the infection has reached and the predictions are made for the total infections. According to the model, the cumulative infections in the USA at the end of the outbreak will be around 4 million. The model prediction of Brazil shows that the peak date is 5 July 2020 and total infections are 3.2 million. The respective predictions for India are 5 August 2020 and 2.4 million. The predictions of India are discussed in the context of lock-down imposed by the government. A broader curve for daily infections against time and a slower decay of the growth rate in the case of India is found to be the signatures of the effect of a strict lock-down. The limitations of the present model are briefly discussed.
References
Batista M (2020) Estimation of the final size of the COVID-19 epidemic. medRxiv preprint. https://doi.org/10.1101/2020.02.16.20023606
Bhardwaj R, Agrawal A (2020) Likelihood of survival of coronavirus in a respiratory droplet deposited on a solid surface. Phys Fluids 32:061704
Quadrat J-P (2020) A simple COVID-19 epidemic model and containment policy in france. medRxiv preprint. https://doi.org/10.1101/2020.04.25.20079434
Ranjan R (2020) Predictions for COVID-19 outbreak in India using epidemiological models. medRxiv preprint. https://doi.org/10.1101/2020.04.02.20051466
Renshaw E (1993) Modelling biological populations in space and time, vol 11. Cambridge University Press, Cambridge
Vattay G (2020) Predicting the ultimate outcome of the COVID-19 outbreak in Italy. arXiv preprint. arXiv:2003.07912
Verma Mahendra K, Ali A, Soumyadeep C (2020) COVID-19 pandemic: power law spread and flattening of the curve. Trans Indian Natl Acad Eng. https://doi.org/10.1007/s41403-020-00104-y
Acknowledgements
The work has been benefited from postings on a online discussion forum of IIT Bombay faculty. I thank Professors Amit Agrawal, E. Chandrasekhar, Amitabh Bhattacharya and Shyamprasad Karagadde for useful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bhardwaj, R. A Predictive Model for the Evolution of COVID-19. Trans Indian Natl. Acad. Eng. 5, 133–140 (2020). https://doi.org/10.1007/s41403-020-00130-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41403-020-00130-w