Manuscript accepted on :
Published online on: --
Plagiarism Check: Yes
Shashank M Patil1, Chandana Kumari V B1, Prithvi S Shirahatti2, Sujay S1, Tejaswini M1, Lakshmi Ranganatha V3, Mallikarjunaswamy C4, Jayanthi M K5 and Ramith Ramu1
1Department of Biotechnology and Bioinformatics, School of Life Sciences, JSS Academy of Higher Education and Research (JSS AHER) Mysuru-570015, Karnataka, India
2Department of Biotechnology, Teresian College, Siddhartha Nagara, Mysuru - 570 011, India
3Department of Chemistry, The National Institute of Engineering, Manandavadi Road, Mysuru, Karnataka, 570008, India
4Department of Chemistry, JSS College of Arts, Commerce and Science, Ooty Road, Mysuru - 5700025, Karnataka, India
5Department of Pharmacology, JSS Medical College, JSS Academy of Higher Education and Research (JSS AHER) Mysuru – 570015, Karnataka, India
Corresponding Author E-mail: ramithramu@jssuni.edu.inDOI : https://dx.doi.org/10.13005/bpj/2033
Abstract
The world has witnessed COVID-19 or SARS-CoV-2 as one of the most hazardous viral outbreak in the history of mankind. Since its emergence in December 2019, it has been affecting the global health with no reported pharmacotherapeutic agent that can neutralize its substantial pathogenicity and escalation around the world. This is attributed to its remarkable molecular pathways followed in course of its life cycle, which is completed in and around the host cell. With the usage of these evolved mechanisms, the virus can effectively invade and replicate in the host cell. The complete analysis of life cycle has resulted in reporting of some molecular targets, which can be neutralised with the usage of pharmacotherapeutic agents. These agents tend to bind to their targets to inactivate them. This review focusses on those targets as well as the potent drugs that currently have been employed to reduce the viral load, in the perspective of its life cycle and pathogenicity. Alongside the drugs that are currently being used, we also report potent drugs that are yet to clear the clinical investigation.
Keywords
COVID-19; Life Cycle; Molecular Targets; Pathogenicity; Pharmacotherapeutics; SARS-CoV-2
Download this article as:| Copy the following to cite this article: Patila S. M, Kumari V. B. C, Shirahatti P. S, Sujay S, Tejaswini M, Ranganatha V. L, Mallikarjunaswamy C, Jayanthi M K, Ramua R. Pharmacotherapy of COVID-19: A Perspective of Pathogenicity and Life Cycle. Biomed Pharmacol J 2020;13(3). |
| Copy the following to cite this URL: Patila S. M, Kumari V. B. C, Shirahatti P. S, Sujay S, Tejaswini M, Ranganatha V. L, Mallikarjunaswamy C, Jayanthi M K, Ramua R. Pharmacotherapy of COVID-19: A Perspective of Pathogenicity and Life Cycle. Biomed Pharmacol J 2020;13(3). Available from: https://bit.ly/3j0RcpZ |
Introduction
The emergence of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) from an animal market in Wuhan city, Hubei province of China in the December of 2019 has created a global health, social and economic crisis, which the world had not witnessed for over a century. It was originally designated as the 2019 novel coronavirus or 2019-nCoV, later as SARS-CoV-2 by the International Committee on Taxonomy of Viruses (ICTV). Subsequently, it was named as coronavirus-19 or COVID-19 by the world health organisation.1,2 To this date, the contagion has spread over 212 countries, resulting in 40,88,848 cases and 2,83,153 deaths, and been officially declared as pandemic by the WHO, owing to its extensive and widespread pathogenicity and enhanced mode of infection compared to its counterparts.3 This has resulted in the world lockdown condition, where international and domestic trades and travels have been called off to avoid further infection and transmission of the disease.4 As a result, financial losses have triggered the economic crisis, with possible fall of 13.0-32.0% global trades and reducing global GDP by 2.0%.5 The literature depicts that symptoms of this virus are like that of pneumonia, including headache, fever, dyspnea, non-productive cough and fatigue. However, patients may also develop acute respiratory distress and hypoxia, which needs timely ventilation.4,5 Transmission of the virus occurs between infected and healthy human individual through inhaling aerosols that are produced by coughing, sneezing and direct close contact.6 Possible animal to human transmissions including bat, snake and pangolin animals are under investigation as the virus is expected to have emerged from the animal market.7 It is supported by the emergence of other two viruses of the same family, middle east respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV-1, which were originated from bats using camels and civet cats as intermediate hosts, respectively.6,7 Thus far, treating the patients is said to be a stressful task as the virus is highly contagious and can affect even the medical personnel. However, timely and efficient supply of personal protective equipment (PPE), detection kits, drugs along with careful treatment can result in the quicker recovery of the patients.8 As the healthcare services largely depend on the supply of PPE and drugs, it becomes essential for the pharmaceutical industries to supply the same. Along with PPE, the important role in controlling the pandemic is played by pharmacotherapeutics.9,10,11 Though vaccines are not yet discovered, it is reported that few of the drugs that are being used for other viruses from the same family have been tested effective and some more are yet to clear the clinical trials against SARS-CoV-2.12 The drugs which were used once to treat MERS-CoV and SARS-CoV-1 are now being employed against SARS-CoV-2, as the latter shares about 80% similar genome with the SARS-CoV-1.6 Many of these drugs possess anti-viral properties and were used to treat viral infections like HIV-1, influenza, Ebola, hepatitis-B and C, and malaria. These are either used individually or in combination based on the response from the patient. Many of these drugs are reported to be used originally as anti-bacterial and antiprotozoal agents, which are also known to possess antiviral properties.9,12 In this review, we report the different drugs used against COVID-19, along with their chemical properties, mode of action, original indication, dosage recommendations, and adverse effects. Apart from drugs being used we also report drugs that are yet to clear their clinical trials. We aim to provide this information to facilitate healthcare providers and people with foundational knowledge on pharmacotherapeutics that are both currently being used and under clinical investigation.
Notable Features OF COVID-19 Pathogenicity
The pathogenicity of COVID-19 is attributed to its structural and functional modifications compared to its counterparts, MERS-CoV and SARS-CoV-1. The SARS-CoV-2 belongs to the genus β-coronavirus, family Coronaviridae and order Nidovirales.6,13 Though several human coronaviruses including HCoV-NL63, HCoV-OC43, HCoV-HKU, and HCoV-229E were reported to circulate in humans for centuries, they were only linked with mild respiratory diseases.14,15 The most lethal variants SARS-CoV-1, MERS-CoV and SARS-CoV-2 have zoonotically been transmitted from other mammals in the recent years.7,16 The most lethal of them, SARS-CoV-2 possesses a large, single-stranded, positive-sense RNA as its genome of approximately 27-32 kb length.17,18 It comprises of 6-10 genes with first gene encoding for replication and transcription. The rest of the genes are structural genes that codes for structural proteins including the spike (S), membrane (M), envelope (E) and nucleocapsid (N) [Figure 1]. Here, membrane constitutes the viral coat and nucleocapsid packages the viral genome. Some of these are transformed into glycoproteins after undergoing glycosylation in the Golgi apparatus.19 The spike (S) is a surface glycoprotein reported to play an important role in binding of the virus to the host cell. The glycoprotein is primed by 2 proteases, Angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2). With ACE2 being responsible for posing as a receptor, SARS-CoV-2 binds to the host cell. Thus, it proves a gateway for the entry of the virus into the human cell.19,20
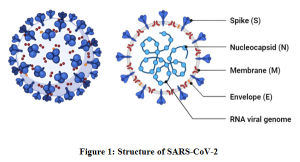 |
Figure 1: Structure of SARS-CoV-2 |
In 2003, the Guangdong province of China reported the outbreak of SARS-CoV-1, leading to acute respiratory distress syndrome (ARDS). Later, the outbreak spread to rest of the world, resulting in 8000 infections and 776 deaths.21,22 Nearly a decade later, Saudi Arabia reported another coronavirus infection in 2012, which was named as MERS-CoV. In this virus, infection begins from a mild respiratory injury leading to severe respiratory disease, and alike SARS-CoV-1, patients witness ARDS and renal failure. This outbreak resulted in more than 2400 infections and 838 deaths.23,24 In December 2019, the SARS-CoV-2 outbreak was witnessed in China, which has reported 90% of the cases till date. These viruses use different animals as their reservoirs with a common origin, bat.25,26 SARS-CoV-2 is also reported to be originated from bats, later believed to be transmitted through pangolin. This is supported by a study, where it was found that a coronavirus isolated from Malayan pangolin possess 99% similarity with SARS-CoV-2; with infected pangolins displaying similar symptoms like that of humans infected with COVID-19.26,27 After all, there is a significant difference in transmission patterns between these viruses. Viral shedding is one of the notable factors where the progeny is released into external environment after completing the reproduction. In case of MERS-CoV and SARS-CoV-1, it occurs only after the onset of symptoms, hence transmission occurs after the medical help.2 However, considering its predominant human-human transmission especially in communities and families, it was reported to spread even far before the occurrence of symptoms and possess similar half-lives (1.1-1.2 hrs) in aerosols.28 Another study reported its extensive transmission, where two proven COVID-19 recovered patients showed positive in tests conducted few days later.29 Meanwhile, the incubation time is also high compared to SARS-CoV-1.30,31 Thus, it takes a great amount of time to show the symptoms before which the infected might spread the disease to several others.32,33 The virus remains viable on solid surfaces like plastic and steel for a considerable amount of time.7 In the perspective of detection and diagnosis, SARS-CoV-2 cannot be detected effectively by RT-PCR, loop-mediated isothermal amplification (LAMP), or chest CT, where SARS-CoV-1 was efficiently diagnosed with RT-PCR alone.34 Owing to these factors, the disease can be declared as more contagious than the other two viruses.
Pharmacotherapeutics Used Against COVID-19
To this date, many of the drugs have been used by medical professionals to treat COVOD-19, either individually or in combination, according to their national healthcare guidelines, though there is no specific drug available to treat the contagion.1,2 However, these drugs are employed due to their proven efficacy against other coronaviruses. Most of them are anti-virals used against diseases like hepatitis, Ebola, influenza, and HIV-1. Some of the drugs with original indications including anti-inflammatory, anti-bacterial, immunomodulatory functions are also used.9,12 These drugs inhibit the viral load at different intervals including host-pathogen interaction, viral entry into the host cell, replication of viral genome and viral polypeptide production [Table 1]. Herein we discuss the effects of these drugs and their mode of action on molecular targets with respect to the viral life cycle.
Table 1: Pharmacotherapeutic agents used against COVID-1911,12,42
| Acting Stage | Classification | Name of the Drug | Chemical Formula
|
Original Indication
|
Possible COVID-19 Indication | Dosage
|
| Viral Entry Inhibitors | Viral S protein Inhibitors | Chloroquine | C18H26ClN3 | Malaria, HIV, Autoimmune Diseases | Off-label use for anti-viral treatment | 500 mg twice for 5 days, orally |
| Hydroxychloroquine | C18H26ClN3O | Malaria, HIV, Autoimmune Diseases | Off-label use for anti-viral treatment | 400 mg on first day followed by 200 mg for four days, twice, orally | ||
| Emodin | C15H10O5 | Polycystic kidney disease | Anti-viral drug under investigation | Unknown | ||
| Promazine | C17H20N2S | Treating psychomotor conditions | Anti-viral drug under investigation | Unknown | ||
| TMPRSS2 Inhibitors | Camostat mesylate | C21H26N4O8S | Chronic pancreatitis | Off‑label use for anti‑viral response | Unknown | |
| K11777 | C32H38N4O4S | Anti-parasitic | Anti-viral drug under investigation | Unknown | ||
| Nafamostat | C19H17N5O2 | Pancreatitis | Off‑label use for anti‑viral response | Unknown | ||
| Hemagglutinin Inhibitors | Nitazoxanide | C12H9N3O5S | Diarrhea | Off-label use for anti-protozoal treatment | Doses for SARS were based on age groups; 1-3-year olds were recommended with 100 mg, 4-11 years with 200 mg, above 12 years with 300 mg for 5 days, orally
|
|
| Arbidol Hydrochloride | C22H26BrClN2O3S | Unknown | Anti-viral drug under investigation | Unknown | ||
| RNA Replication Inhibitors | Nucleoside Analogs | Remdesivir | C27H35N6O8P | Ebola virus, MERS-CoV | Off-label use for anti-viral treatment | 200 mg on first day, followed by 100 mg for up to 10 days intravenously |
| Ribavirin | C8H12N4O5 | Hepatitis-A, Hepatitis-B, SARS | Off-label use for anti-viral treatment | 400 mg for 14 days, twice a day | ||
| Favipiravir | C5H4FN3O2 | Influenza | Off-label use for anti-viral treatment | Unknown | ||
| Ivermectin | C48H74O14 | Unknown | Off-label use for HIV-1 and anti-viral treatment | Unknown | ||
| Sofosbuvir | C22H29FN3O9P | Hepatitis C virus infection | Off‑label use for anti‑viral treatment | Unknown | ||
| Protein Synthesis Inhibitors | Protease Inhibitors | Lopinavir and Ritonavir | C74H96N10O10S2 | HIV-1 | Off-label use for HIV-1 treatment | 200 mg -100 mg for 14 days through oral consumption |
| Nelfinavir | C33H49N3O7S2 | HIV-1 | Off-label use for HIV-1 and anti-viral treatment | Unknown | ||
| Neuraminidase Inhibitors | Oseltamivir | C16H28N2O4 | Influenza | Off‑label use for anti‑viral treatment | Unknown | |
| Zanamivir | C12H20N4O7 | Influenza | Off‑label use for anti‑viral treatment | Unknown | ||
| Unknown
|
Bactericidal | Azithromycin | C38H72N2O12 | Bacterial Infections | Antibiotic | Unknown |
Drugs Inhibiting The Viral Adsorption
The mode of infection of COVID-19 chiefly begins with inhaling aerosols from infected person, which then enters the respiratory system like that of pneumonia and tuberculosis.4 These aerosols comprising of viable viral particles enter the alveolar cavity and unload the virus thus letting the virus gain entry into the alveolar cells. Apart from alveolar cells, the virus is reported to affect heart cells, as the respiratory system interact with the heart directly. This is supported by the higher amount of secretion of ACE2, which aids in the binding of virus to the host cell. As the heart is said to be the principle organ for circulation, the virus spreads throughout the body resulting in the damage of digestive system. The occurrence of nausea, vomiting and diarrhea supports this hypothesis. Further, the virus is reported to damage kidney and liver, which is supported by the symptoms showing renal failure and bile duct, respectively.35 It is worth mentioning about spike (S) protein, which facilitates the entry of the virus. It is primed by the host cell protease and recognized by the cellular receptor. In case of humans, a serine protease receptor known as TMPRSS2 is responsible for the priming the spike protein and ACE2 is employed as a receptor.20,36 In other words, the surface glycoprotein plays the role of a key to open the door and facilitate the viral entry into the host cell. It is also reported to show high binding affinity with ACE2, compared to SARS-COV-1. Based on the cryo-EM structure, it is evident that out of several functional domains the receptor binding protein (RBD), fusion domain (FD) and the S2 cleavage prove to be potential targets for drug development.37,38
Chloroquine
Chloroquine and its analogues have been used to control malaria since early 1900s.39 The multimodal properties of chloroquine analogues and their minimal toxicity profile has initiated clinical investigation to prove their effect on virus-induced illnesses such as HIV-associated immune reconstitution inflammatory syndrome.40 Chloroquine is reported to impair the glycosylation process of ACE2, thus impairing its interaction with spike protein, subsequently the host cell-virus interaction.39 These analogues also increase the pH level and thus inhibit activities like protein biosynthesis and dysfunction of enzymes. These mechanisms of chloroquine-mediated pH alteration are been applied to disintegrate viral replication. Since the mechanism proves to be potent to treat COVID-19, it has been used alongside the other drugs.40 For treating COVID-19, 500mg of the drug is given twice a day for 10 days.42 In February 2020, a study reported the inhibitory activity of chloroquine against SARS-CoV-2 with an EC50 at 48 hours of 1.13 µM in Vero E6 cells.43 These findings have upheld the clinical usage of chloroquine in China at the time of outbreak.44 Results from trials revealed that chloroquine inhibits the exacerbation of COVID-19. However, high doses of chloroquine may result in the development of retinopathy, though it is well tolerated with some common adverse effects like nausea, vomiting, abdominal cramps, and metallic taste.45
Hydroxychloroquine
Hydroxychloroquine also belongs to the analogues of chloroquine and has been treated as one of the most effective therapeutic option for the management of COVID-19.46 Both chloroquine and hydroxychloroquine prevent clathrin complex formation during endocytosis. Besides, hydroxychloroquine uses the same mechanism of chloroquine; increasing the intracellular pH level to disintegrate the cellular activities of the virus [Figure 2]. In addition to this, hydroxychloroquine is also reported to impair the glycosylation of ACE2 protein, thus inhibiting the host cell-virus adsorption through impairing the interaction of spike protein.39
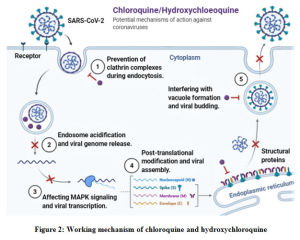 |
Figure 2: Working mechanism of chloroquine and hydroxychloroquine |
Hydroxychloroquine was assessed using pharmacology based pharmacokinetic (PBPK) modelling on Vero cells against SARS-CoV-2 along with chloroquine. This study revealed that potency of hydroxychloroquine (EC50 of 0.72 µM) was greater than that of chloroquine (EC50 of 5.47 µM).46 In addition to this, a similar open-labelled, non-randomized study conducted in France showed positive effect against COVID-19 in combination with azithromycin.47 In the wake of this evidence, U.S. FDA cleared this drug for emergency uses across USA.48 Though dosing recommendations ae not yet deduced, current pharmacokinetic models suggest 400 mg of the drug twice on the first day, followed by 200 mg twice a day for four days, orally.46 It has been reported that hydroxychloroquine possesses superior cytotoxic effects compared to chloroquine.49 These include dermatologic adverse effects like lichenoid reactions, photosensitivity, and acute generalized exanthematous pustulosis can occur with hydroxychloroquine. It has also been reported that persons with history of diabetes showed hypoglycemic symptoms and loss of consciousness upon consumption.50 In addition to chloroquine and hydroxychloroquine, there are two more drugs that work using the same mechanism of impairing ACE2 function, known as emodin and promazine. The original indication of emodin was found to be under investigation against the polycystic kidney disease. It is yet to clear the clinical trials as it is still being investigated for its anti-viral effects. Promazine is a discontinued drug used for treating psychomotor conditions. It is possible COVID-19 is off-label use anti-viral response. The dosage information and adverse effects on COVID-19 patients of these two drugs is not yet revealed, as they are still being investigated.12
Like ACE2, another enzyme known as TMPRSS2 also facilitates the viral entry to the cell. When the COVID-19 virus tends to enter the host cell, it becomes essential for the spike protein to get cleaved and activated to facilitate the viral entry to the cell.51 Therefore, inhibition of the TMPRSS2 protease would become a potential target for anti-viral drug development. Recently three drugs known as camostat mesylate, nafamostat and K11777 have shown to possess potent effects against COVID-19. Camostat mesylate was originally designed to treat chronic pancreatitis.12 Whereas, K11777 was used as an anti-parasitic agent, to treat toxoplasmosis.52 Nafamostat was originally used as an anti-microbial and anti-coagulant agent.53,54 Though all three drugs are potent to inhibit TMPRSS2, their clinical evaluation is yet to be completed. Hence, their dosage recommendations and adverse effects are not revealed yet.
Hemagglutinin esterases are the group of viral envelope glycoproteins that facilitate the surface adsorption of virus and the host cell membrane.36 According to a study conducted by Zeng et al, the CoV-hemagglutinin (CoV-HE) complex underwent considerable modifications to facilitate the optimal binding of the virus into the host cell. It is also reported that, the plasticity of CoV-HE is attributed to the functional redundancy between spike protein and hemagglutinin component.55 Thus, it becomes evident that hemagglutinin also play an important role in the binding of the virus to the host cell, as well as provide another potential site for drug development. Aiming the exploitation of this mechanism, few drugs have been employed to treat COVID-19.
Nitazoxanide
Nitazoxanide is a U.S FDA approved benzamide drug, which is used to treat diarrhea caused by Giardia spp.42 It is known for its anti-protozoal activity and has also been used against various Gram-positive and Gram-negative bacteria.56,57 Another reported study of nitazoxanide reveals it also shows anti-viral activity, against influenza.58 After ingestion, it gets converted into its active state metabolite, known as tizoxanide. It inhibits the hemagglutinin formation and protein implantation in plasma membrane.59,60 It may facilitate IFN-1 production to inhibit hemagglutinin, thus showing its anti-viral effects.61 The pharmacological effects of nitazoxanide proved to be effective against SARS-CoV-2 with EC50 value of 2.12 µM in Vero E6 cells at 48 hours of incubation.62 But in case of human trials, a study conducted on 5 positive patients proved its inefficiency. Sub-group analysis of these patients showed no difference even after days of hospitalization. Doses recommended for SARS were based on age groups; 1-3-year olds were recommended with 100 mg, 4-11 years with 200 mg, above 12 years with 300 mg for 5 days, orally. The side effects of nitazoxanide include, abdominal cramps, nausea, headache along with discoloration of urine, diarrhea, dizziness and urticaria.63 In addition to nitazoxanide, another potent entry inhibitor known as arbidol hydrochloride or umifenovir also works using the same mechanism. The drug has been approved by China and Russia. Arbidol is reported to prevent the fusion of the viral envelope with endosome after endocytosis. The recommended dosage for COVID-19 and adverse effects on patients are not yet revealed as the drug is yet to clear the trials.12,64
Drugs Inhibiting the Viral Genome Replication
After the successful entry of the virus into the host cell, the virus replicates its genome irrespective of its nature. In case of COVID-19, as the genome is RNA, the replication is done using both continuous and discontinuous mechanisms at cytoplasmic membranes. The replication process is reported to be mediated by 20-kb length replicase gene. The replicase complex is composed of 16-subunits and cellular proteins. Apart from RNA-dependent RNA polymerase and RNA helicase, COVID-19 is believed to possess different RNA processing enzymes including endoribonuclease, ADP ribose 1’-phosphatase, and 2’-O-ribose methyl transferase. These proteins are present at cell membrane and aid in the production of mature RNA in the cytoplasmic membrane.65 The impairment of replication can prove a potential target for few drugs to inhibit the viral replication.
Remdesivir
Remdesivir is an anti-viral drug that was originally developed to treat Ebola hemorrhagic fever with ongoing trials. It is an investigational monophosphoramidate of an adenosine analog developed by Gilead Sciences, Inc.66,67 Though it has not been approved for usage globally, remdesivir is being used to treat COVID-19 with phase 3 clinical trials on the run.68 Both in vitro and in vivo studies were conducted and revealed that remdesivir has the antiviral activity against filoviridae, paramyxoviridae and the coronaviridae including MERS-CoV, SARS-CoV-1 and SARS-CoV-2.69,70 It has been reported that remdesivir has shown an EC50 at 48 hours of 0.77 µM in Vero E6 cells.43 Being a pro-drug, remdesivir gets metabolized in cells and tissues to become an active nucleoside triphosphate that interferes with RNA polymerase activity of the virus [Figure 3]. Other potential mechanisms like lethal mutations and chain termination may also pose beneficial to use it as an anti-viral drug.71 It is reported that once remdesivir was used to treat Ebola patients.67 Another study conducted by Sheahan et al., revealed its efficiency against MERS-CoV and SARS-CoV-1 activity.69 However, the dose under clinical investigation is 200 mg on first day, followed by 100 mg for up to 10 days intravenously.62 The first report of treating COVID-19 patient came in US, where a patient received the treatment with remdesivir for 7 days and found to have shown clinical improvement, though nasopharyngeal swab was positive.72 This study reported no side effects but, few cases revealed minor adverse effects like nausea, vomiting and rectal bleeding.62
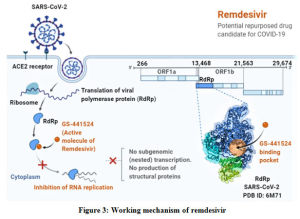 |
Figure 3: Working mechanism of remdesivir |
Ribavirin
Ribavirin affects the viral RNA synthesis, thus disintegrates the viral life cycle as it belongs to the class of purine nucleoside analog. Being a prodrug, it gets modified in liver upon consumption, where its metabolic structure starts mimicking with guanosine. This enables its incorporation into viral RNA, ultimately resulting in the RNA synthesis inhibition.73 As the molecular mimicry can occur in many viruses with the presence of RNA, Ribavirin can be found effective against an array of viruses including hepatitis-B, hepatitis-C, and respiratory syncytial virus. This stipulated the investigation for anti-viral activity of the drug against SARS and MERS outbreaks.74 During the SARS outbreak, 70% of hemolysis and 49% of anemia cases were reported out of 126 treated with ribavirin.75,76 US had to restrict the use of ribavirin due to lack of in vitro susceptibility and high toxicity levels. With respect to SARS-CoV-2, Wang et al., assessed it is in vitro activity and found an EC50 of 109.5 µM, which is nearly 100 times less potent compared to remdesivir.76 These findings initiated the usage of ribavirin on COVID-19 and it is now recommended that oral ribavirin should be given as 400 mg for 14 days, twice a day. However, adverse effects may arise in few patients including hemolytic anemia, hypomagnesemia and hypocalcemia. Pregnant women are advised not to use ribavirin because of its embryonic toxicity.74
Favipiravir
Favipiravir was developed by a Japanese firm known as Toyoma Chemicals. It acts as an RNA polymerase inhibitor by the mechanism of molecular mimicry. It structurally resembles guanine, and through competitive inhibition, reduces the efficacy of viral replication like remdesivir.77 Though reports state its pharmacological potency against SARS-CoV-2, and better pharmacological activity than arbidol hydrochloride1, favipiravir has got less preclinical support.11,42 However, it can be used against influenza, the disease it was formulated to treat. Clinical studies have been on the run, where favipiravir is used in combination with IFN-α to assess viral inhibition and immune system enhancement. National Medical Products Administration of China approved favipiravir as the first anti-COVID-19 drug in China, as it successfully cleared the clinical tests with minimal side effects.9 Sofosbuvir, which was used against hepatitis-c infection, uses the same molecular mechanism to impair viral genome replication. Its dosage and side effects are not yet deciphered as the drug is still under clinical investigation.12
Ivermectin
Ivermectin is an anti-parasitic drug approved by U.S FDA which has got the potency to show anti-viral properties. It has been used to treat HIV-1 and dengue viruses. The mechanism of ivermectin reveals that it can dissociate the preformed IMPα.β1 heterodimer, which aids in the protein displacement.78 As the protein displacement is essential for the maintenance of viral replication, targeting the protein displacement across the host cell would be a feasible option to inhibit viral life cycle. A recently conducted study reported that ivermectin reduced the viral RNA up to 5000-fold after infection with SARS-CoV-2 for 48 hours.9,79,80 With the proven results for anti-parasitic activity, ivermectin is under clinical trials to prove its potency against COVID-19.
Drugs Inhibiting the Viral Protein Synthesis
Once the replication of genome becomes complete, the translation starts in the cytoplasm. Polyproteins and structural proteins are produced because of translation. Two polypeptides known as pp1a and pp1ab are processed by either a chymotrypsin-like protease (3CLpro) or main protease (Mpro) and one or two papain-like proteases into 16 nsps. sgRNA’s of virus get translated into produce all the structural and accessory proteins. Here, it becomes worthwhile to mention the production all the four main structural proteins, including spike protein.65 Thus, the protein production can be targeted to inhibit the viral growth, as it involves a set of enzymes posing as potential targets.
Lopinavir and Ritonavir
Lopinavir is reported to be an aspartic acid protease inhibitor developed to treat human immunodeficiency virus-1 (HIV-1) [Figure 4]. It is admistered along with ritonavir in a fixed-dose combination.81 Ritonavir is a CYP3A4 and CYP450 inhibitor that enhances the concentration, half-life as well as the pharmacokinetic activity of lopinavir. It has been revealed that the combination of lopinavir and ritonavir provides potent and sustained reduction of viral activities in HIV-1 patients.82,83 This has prompted interest in the assessment of the efficacy of the drug of other viruses including COVID-19. Studies have revealed its efficacy on both SARS and MERS viruses.84 In addition to this, Chu et al., reported that 4mg/mL of lopinavir and 50 mg/mL of ribavirin inhibited SARS-CoV-1 after 48 hours incubation.82 Similarly, another study conducted by de Wilde et al., revealed that lopinavir inhibited SARS-CoV-1 with EC50 17.1 ± 1 in Vero cells.83 They reported that combination of lopinavir and ritonavir possessed better anti-viral activity than lopinavir alone. Thus, the results became a pavement for the treatment of SARS-CoV-1. In case of COVID-19, it proves to be efficient in case of mild and moderate infections.12 But another clinical study found no benefits of lopinavir and ritonavir in case of severe infections. Lopinavir/ritonavir is formulated as a single tablet and a dose 200 mg -100 mg for 14 days through oral consumption is advised.81 Adverse effects of lopinavir include diarrhea, fatal pancreatitis, and hepatic decomposition.85
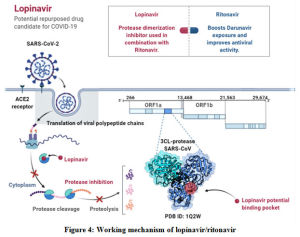 |
Figure 4: Working mechanism of lopinavir/ritonavir |
Nelfinavir
Nelfinavir was originally developed for treating HIV-1 in combination with other anti-virals. It acts as a HIV-1 protease inhibitor, inhibiting the life cycle viral life cycle. It was used with other drugs via alternate mechanisms. But with the development of newer drugs for HIV, nelfinavir was excluded from the list. It is reported that the drug binds to the active site of HIV-1 protease enzyme and inhibits the cleavage of precursors of Gag-Pol polyprotein chain that are essential for the survival of HIV-1 inside the host. The residues left after the molecular process are no longer infectious. Nelfinavir was proved a potent drug against SARS during the 2002 outbreak, with in vitro approaches.86 In this study, it successfully inhibited the SARS-CoV-1 in Vero cells. Again, it was used against SARS-CoV-2 because of the >80% similar sequence with SARS-CoV-1.21 Out of the 20 drugs assessed, nelfinavir proved the most efficient drug. As all these studies were conducted on cell lines, human dose for COVID-19 is still unknown. However, for patients with hepatic maladies, nelfinavir is not recommended.87,88 Major side effects of nelfinavir include nausea, flatulence and diarrhea.89
Neuraminidase determines the transmissibility of the influenza virus by interacting with sialic acid to cleave it, which in turn helps the virus replication and shedding. In fact, it facilitates the viral breakout from the host cell after the completion of viral genome replication, translation, and assembly of the viral particles.90 Oseltamivir and zanamivir are the two more drugs reported to have their effects on viral genome translation. Both these drugs were used against influenza, by targeting a surface glycoprotein known as neuraminidase. These neuraminidase inhibitors are yet to clear the clinical trials. Therefore, the details on dosage and adverse effects are currently not revealed.12 Apart from drugs with anti-viral indication, azithromycin is the only drug reported to have anti-bacterial effects as its original indication, though employed against COVID-19. It is reported that, azithromycin was originally designed for the prevention of secondary bacterial infection and it also possesses anti-viral activity.12,91 Damle et al., have reported the 3 possible mechanisms shown by azithromycin to bring out the reduction in viral load. The first mechanism is like chloroquine and hydroxychloroquine, increasing the intracellular pH level of the cell to dismantle the viral replication process to halt the replication. The second mechanism suggests that the viral load reduction is attributed to the ability of azithromycin to induce pattern recognition receptors (IFNs, and IFN-stimulated genes). In addition to these, it is reported to act directly on the bronchial epithelial cells to regulate their normal function by reducing mucus secretion.91The dosage recommendations of azithromycin are 500 mg on first day, followed by 250 mg 4 days.12
Challenges in Drug and Development
To this date, there is no reported drug that completely inhibits the viral activity in case of COVID-19. In the drug development, it becomes essential to have animal models to test and reveal the mechanisms underlying COVID-19 pathogenicity.85 Though cell cultures prove to be efficient animal, models give better results in case of physiological analysis.92 As most of the research has been conducted with the COVID-19 potent therapeutics is linked with Vero cells,11 the results may vary in case of human beings. In a study, various animal models tested for SARS-CoV replication, showed significant infection.93 Tests against MERS-CoV, they were not vulnerable to the infection due to the absence of DPP4 receptor.94 But in case of SARS-CoV-1 they showed up with severe disease symptom.95 Therefore, development of suitable animal models to conduct pharmacotherapeutic studies in order to understand the behaviour of potential targets.
Apart from the requirement of animal models, clinical dosage of the drugs also plays an important role in the COVID-19 therapy. To this date, no drug has been available with international recommendations.96 All the above reported drug dosages are given either under national healthcare guidelines or are still under clinical investigation. Besides, due to their variety of original indications, these drugs cannot be used directly against COVID-19.9,12 Furthermore, physiological changes must be considered, which arise because of the drug action. Cytotoxicity is one of the major assays performed in case of drug development as it determines the viability of cells against the developed drug. Being popular drugs used to treat COVID-19, both chloroquine and hydroxychloroquine are reported to cause ventricular dysrhythmias, hyperkalemia and systemic lupus erythematosus (SLE) respectively, in case of abnormal dosages.49 In addition to this, many of the drugs have not cleared QT analysis, which is essential for the normal functioning of the ECG, including hydroxychloroquine. Considering these factors, the need to identify a potential drug attribute has been the need of the hour.97,98
Alongside the dosage aspects, treating COVID-19 patients with non-COVID-19 diseases/disorders becomes hectic due to the drug-drug reaction between COVID-19 and non-COVID-19 therapeutic agents. As a result, patients who can be successfully tested for drug dosage against COVID-19 cannot be tested any longer for other drug because of the effect of other non-COVID-19 therapeutic agent.99 For example, if a COVID-19 positive patient with additional rare genetic disease or diabetes may witness adverse effects due to the administration of both type of drugs.100 Therefore, it becomes difficult to analyse accurately the pharmacological effect of the COVID-19 drug in the presence of non-COVID-19 drug. Thus, patients may also hamper the R&D activities associated with pharmacotherapy. Despite the reduction in the spreading of the disease, the quarantine and lockdown conditions have also resulted in handicapping the R&D activities like clinical data investigation including case studies, patient visits, data on pharmacokinetic, pharmacodynamic, efficacy limits, cytotoxicity limits and evidences.101 Though clinicians have effectively handled these issues with the help of technology and taken umpteen care towards collection and handling of samples from remote areas, careful interaction with patients, effective usage of digital media to cover data collection, assessment and presentation there exists several gaps owing to which the containment of the disease has not been achieved. Quantitative analysis has been reduced due to altered schedules of pharmacokinetic and pharmacodynamic data collections, and other model-based analysis. This has significantly affected the vaccine production, from which the COVID-19 would have been controlled at a notable.101,102
Conclusion
The outbreak of COVID-19 has raised unprecedented challenges before the world relating to healthcare and drug development. Pharmaceutical industry has responded to the situation with maximal efforts and still it is being employed to it. Through much of the research has been conducted out on the life cycle and pathogenesis, most of the potential targets to tackle have been revealed. These targets exist at different stages of life cycle of the virus and can be effectively neutralised with accurate and adequate dose of specific drugs. As the review highlights about the different drugs that are being employed to treat the COVID-19, it specifically denotes the mechanisms of both the virus and the drug, their point of contact, and the pathogenicity at molecular level. Though many of the drugs are being used against the pandemic, their universal dosage levels and other pharmacological details are not revealed as they are still given under national healthcare guidelines of respective countries. To this date, WHO has not recommended any of the above reported drug although a few of them have received approval by the FDA. Thus, it becomes evident that advanced research on the drugs is to be done. It is worthwhile to highlight about using animal models to observe effective results. With the animal models, the complete pathophysiology of the disease as well as the effect of the drug can be understood. Also, more potent drugs should be tried with clinical investigations to report their efficacy against the COVID-19; along with the combination of drugs with different dosages against induced COVID-19 models. The best results can be obtained with the pharmaceutical industry employing ‘One Health’ approach where comprehensive efforts from different sectors sharing the data to effectively control the pandemic.
References
- Patil SM, Kumari VC, Shirahatti PS, et al. COVID-19 infection: The prospects of pharmacotherapy. Int. J. Health. Allied. Sci. 9(5): 111-113 (2020).
- Kumari VC, Patil SM, Shirahatti PS, et al. The current status and perspective for the emerging pandemic: COVID-19. Int. J. Pharm. Pharm. Sci. 12(8): 1-10 (2020).
- Grossman S, Sandhu P, Sproat C, Patel V. Provision of dental services at a single institution in the UK’s epicentre during the COVID-19 pandemic. Br Dent J. 228(12): 964-970 (2020)
- Taghrir MH, Akbarialiabad H, Marzaleh MA. Efficacy of mass quarantine as leverage of health system governance during COVID-19 outbreak: a mini policy review. Arch Iran Med. 23(4): 265-267 (2020).
- Ogbolosingha AJ, Singh A. COVID-19 pandemic: Review of impediments to public health measures in Sub-Saharan Africa. Am J Prev Med. 6(3): 68-75 (2020).
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395(10223): 497-506 (2020).
- Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 395(10223): 514-23 (2020).
- Shereen MA, Khan S, Kazmi A, et al. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv.Res. 24: 91-98 (2020).
- Nadeem MS, Zamzami MA, Choudhry H, et al. Origin, potential therapeutic targets and treatment for coronavirus disease (COVID-19). Pathogens. 9(4): 307 (2020).
- Cook TM. Personal protective equipment during the coronavirus disease (COVID) 2019 pandemic–a narrative review. Anaesthesia. 75(7): 920-927 (2020).
- Tu Y-F, Chien C-S, Yarmishyn AA, et al. A review of SARS-CoV-2 and the ongoing clinical trials. Int J Mol Sci. 21(7): 2657 (2020).
- Pang J, Wang MX, Ang IYH, et al. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J Clin Med. 9(3): 623 (2020).
- McCreary EK, Pogue JM. Coronavirus Disease 2019 Treatment: A Review of Early and Emerging Options. Open. Forum. Infect. Dis. 7(4): ofaa105 (2020).
- Shetty R, Ghosh A, Honavar SG, Khamar P, Sethu S. Therapeutic opportunities to manage COVID-19/SARS-CoV-2 infection: Present and future. Indian J Ophthalmol. 68(5): 693 (2020).
- Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int. J. Antimicrob. Agents. 55(5):105951 (2020).
- Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17(3): 181-192 (2019).
- Ye Z-W, Yuan S, Yuen K-S, Fung S-Y, Chan C-P, Jin D-Y. Zoonotic origins of human coronaviruses. Int J Biol Sci. 16(10): 1686 (2020).
- Raoult D, Zumla A, Locatelli F, Ippolito G, Kroemer G. Coronavirus infections: Epidemiological, clinical and immunological features and hypotheses. Cell. Stress. 4(4): 66 (2020).
- Smith EC, Denison MR. Coronaviruses as DNA wannabes: a new model for the regulation of RNA virus replication fidelity. PLoS. Pathog. 9(12): e1003760. (2013).
- Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA. Biol. 8(2): 270-279 (2011).
- De Wit E, Van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14(8): 523 (2016).
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181(2): 271-280.e8 (2020).
- Xu R-H, He J-F, Evans MR, et al. Epidemiologic clues to SARS origin in China. Emerg. Infect. Dis. 10(6):1030 (2004).
- Peiris JSM, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat. Med. 10(12): S88-S97, (2004).
- Memish ZA, Mishra N, Olival KJ, et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 19(11): 1819 (2013).
- Rahman A, Sarkar A. Risk factors for fatal middle east respiratory syndrome coronavirus infections in Saudi Arabia: analysis of the WHO Line List, 2013–2018. Am. J. Public. Health. 109(9): 1288-1293 (2019)
- Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579(7798): 270-273 (2020).
- Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive. Care. Med. 46(4): 586-590 (2020).
- Xiao K, Zhai J, Feng Y, et al. Isolation and characterization of 2019-nCoV-like coronavirus from Malayan pangolins. Nature. 583: 286–289 (2020).
- Van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 382(16): 1564-1567 (2020).
- Lan L, Xu D, Ye G, et al. Positive RT-PCR test results in patients recovered from COVID-19. Jama. 323(15): 1502-1503 (2020).
- Lessler J, Reich NG, Brookmeyer R, Perl TM, Nelson KE, Cummings DAT. Incubation periods of acute respiratory viral infections: a systematic review. Lancet. Infect. Dis. 9(5): 291-300 (2009).
- Jiang X, Rayner S, Luo M. Does SARS‐CoV‐2 has a longer incubation period than SARS and MERS? J. Med. Virol. 92(5): 476-478 (2020).
- Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. A novel coronavirus emerging in China—key questions for impact assessment. N. Engl. J. Med. 382(8): 692-694 (2020).
- Nuttall I, Dye C. The SARS wake-up call. Science (80- ). 339(6125): 1287-1288 (2013).
- Yu L, Wu S, Hao X, et al. Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform. Clin. Chem. 66(7):975-977 (2020).
- Li B, Yang J, Zhao F, et al. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin. Res. Cardiol. 109(5): 531-538 (2020)
- Banerjee N, Mukhopadhyay S. Viral glycoproteins: biological role and application in diagnosis. Virusdisease. 27(1): 1-11 (2016).
- Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94(7):e00127-20 (2020).
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80- ). 367(6483): 1260-1263 (2020).
- Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: an old drug against today’s diseases. Lancet. Infect. Dis. 3(11): 722-727 (2003).
- Savarino A, Gennero L, Sperber K, Boelaert JR. The anti-HIV-1 activity of chloroquine. J. Clin. Virol. 20(3): 131-135 (2001).
- Vincent MJ, Bergeron E, Benjannet S, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2(1): 1-10 (2005).
- Barlow A, Landolf KM, Barlow B, et al. Review of emerging pharmacotherapy for the treatment of coronavirus disease 2019. Pharmacother. J. Hum. Pharmacol. Drug. Ther. 40(5): 416-437 (2020)
- Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell. Res. 30(3): 269-271 (2020).
- Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 55(4): 105932 (2020).
- Khuroo MS, Sofi AA, and Khuroo M. Chloroquine and Hydroxychloroquine in Coronavirus Disease 2019 (COVID-19). Facts, Fiction & the Hype. A Critical Appraisal. Int. J. Antimicrob. Agents. 56(3): 106101 (2020).
- Yao X, Ye F, Zhang M, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 71(15):732–739 (2020).
- Gautret P, Lagier J-C, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 56(1): 105949. (2020).
- Molina JM, Delaugerre C, Le Goff J, et al. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Mal. Infect. 50(4): 382-387 (2020).
- Chary MA, Barbuto AF, Izadmehr S, Hayes BD, Burns MM. COVID-19: Therapeutics and their toxicities. J. Med. Toxicol. 16(3): 10-1007 (2020).
- Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. Jama. 323(18): 1824-1836 (2020).
- Glowacka I, Bertram S, Müller MA, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85(9): 4122-4134 (2011).
- Chaparro JD, Cheng T, Tran UP, et al. Two key cathepsins, TgCPB and TgCPL, are targeted by the vinyl sulfone inhibitor K11777 in in vitro and in vivo models of toxoplasmosis. PLoS. One. 13(3): e0193982 (2018).
- Inman RD, Chiu B. Nafamostat mesylate, a serine protease inhibitor, demonstrates novel antimicrobial properties and effectiveness in Chlamydia-induced arthritis. Arthritis. Res. Ther. 14(3): R150 (2012).
- Lee ST, Cho H. The Use of Nafamostat Mesilate as an Anticoagulant during Continuous Renal Replacement Therapy for Children with a High Risk of Bleeding. J. Korean. Soc. Pediatr. Nephrol. 18(2): 98-105 (2014).
- Zeng Q, Langereis MA, Van Vliet ALW, Huizinga EG, De Groot RJ. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. 105(26): 9065-9069 (2008).
- Amadi B, Mwiya M, Musuku J, et al. Effect of nitazoxanide on morbidity and mortality in Zambian children with cryptosporidiosis: a randomised controlled trial. Lancet. 360(9343): 1375-1380 (2002).
- Finegold SM, Molitoris D, Väisänen M-L. Study of the in vitro activities of rifaximin and comparator agents against 536 anaerobic intestinal bacteria from the perspective of potential utility in pathology involving bowel flora. Antimicrob. Agents. Chemother. 53(1): 281-286 (2009).
- Rossignol J-F. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antiviral. Res. 110: 94-103 (2014).
- Rossignol J-F, Stachulski A V. Syntheses and antibacterial activities of tizoxanide, an N-(nitrothiazolyl) salicylamide, and its O-aryl glucuronide. J. Chem. Res. 23(1): 44-45 (1999).
- Rossignol JF, La Frazia S, Chiappa L, Ciucci A, Santoro MG. Thiazolides, a new class of anti-influenza molecules targeting viral hemagglutinin at the post-translational level. J. Biol. Chem. 284(43): 29798-29808 (2009).
- Clerici M, Trabattoni D, Pacei M, Biasin M, Rossignol J-F. The anti-infective Nitazoxanide shows strong immumodulating effects. J. Immun. 186(1): 155.21 (2011).
- Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 395(10236): 1569-1578 (2020).
- Gamiño-Arroyo AE, Guerrero ML, McCarthy S, et al. Efficacy and safety of nitazoxanide in addition to standard of care for the treatment of severe acute respiratory illness. Clin. Infect. Dis. 69(11): 1903-1911 (2019).
- Kadam RU, Wilson IA. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. 114(2): 206-214 (2017).
- Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. S1684-1182(20) 30082-7 (2020).
- Siegel D, Hui HC, Doerffler E, et al. Discovery and Synthesis of a Phosphoramidate Prodrug of a Pyrrolo [2, 1-f][triazin-4-amino] Adenine C-Nucleoside (GS-5734) for the Treatment of Ebola and Emerging Viruses. J. Med. Chem. 60(5) 1648–1661 (2017).
- Mulangu S, Dodd LE, Davey RT Jr, et al. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 381(24): 2293-2303 (2019).
- Hoenen T, Groseth A, Feldmann H. Therapeutic strategies to target the Ebola virus life cycle. Nat. Rev. Microbiol. 17(10): 593-606 (2019).
- Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9(396): eaal3653 (2017).
- Lo MK, Feldmann F, Gary JM, et al. Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci. Transl. Med. 11(494): eaau9242 (2019).
- Agostini ML, Andres EL, Sims AC, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 9(2):e00221-18 (2018).
- Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382(10): 929-936. (2020).
- Crotty S, Cameron C, and Andino R. Ribavirin’s antiviral mechanism of action: lethal mutagenesis? J. Mol. Med. 80(2): 86-95 (2002).
- Knowles SR, Phillips EJ, Dresser L, Matukas L. Common adverse events associated with the use of ribavirin for severe acute respiratory syndrome in Canada. Clin. Infect. Dis. 37(8): 1139-1142 (2003).
- Tan ELC, Ooi EE, Lin C-Y, et al. Inhibition of SARS coronavirus infection in vitro with clinically approved antiviral drugs. Emerg. Infect. Dis. 10(4): 581 (2004).
- Booth CM, Matukas LM, Tomlinson GA, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. Jama. 289(21):2801-2809 (2003).
- Furuta Y, Komeno T, Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Japan. Acad. Ser. B. 93(7):449-463 (2017).
- Wagstaff KM, Sivakumaran H, Heaton SM, et al. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 443(3): 851-856 (2012).
- Caly L, Wagstaff KM, Jans DA. Nuclear trafficking of proteins from RNA viruses: potential target for antivirals? Antiviral. Res. 95(3): 202-206 (2012).
- Yang P, Wang X. COVID-19: a new challenge for human beings. Cell. Mol. Immunol. 17(5): 555-557 (2020).
- Cvetkovic RS, Goa KL. Lopinavir/ritonavir: a review of its use in the management of HIV infection. Drugs. 63(8): 769-802 (2003).
- Chu CM, Cheng VCC, Hung IFN, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 59(3): 252-256 (2004).
- De Wilde AH, Jochmans D, Posthuma CC, et al. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents. Chemother. 58(8): 4875-84 (2014).
- Cao B, Wang Y, Wen D, et al. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 382: 1787-1799 (2020).
- Adachi A, Miura T. Animal model studies on viral infections. Front. Microbiol. 5: 672 (2014).
- Yamamoto N, Yang R, Yoshinaka Y, et al. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 318(3): 719–725 (2004).
- Trapé M, Barnosky S. Nelfinavir in Expanded Postexposure Prophylaxis Causing Acute Hepatitis With Cholestatic Features TWo Case Reports. Infect. Control. Hosp. Epidemiol. 22(6): 333-334 (2001).
- Khaliq1 Yasmin 2, Gallicano K, Seguin I, et al. Single and multiple dose pharmacokinetics of nelfinavir and CYP2C19 activity in human immunodeficiency virus‐infected patients with chronic liver disease. Br. J. Clin. Pharmacol. 50(2): 108-115 (2000).
- Miller CD, El‐Kholi R, Faragon JJ, Lodise TP. Prevalence and risk factors for clinically significant drug interactions with antiretroviral therapy. Pharmacother. J. Hum. Pharmacol. Drug. Ther. 27(10): 1379-1386 (2007).
- Benton A. Ebola at a distance: a pathographic account of anthropology’s relevance. Anthropol. Q. 90(2): 495-524 (2017).
- Damle B, Vourvahis M, Wang E, et al. Clinical Pharmacology Perspectives on the Antiviral Activity of Azithromycin and Use in COVID-19. Clin. Pharmacol. Ther. 108(2): 201-211 (2020).
- Barré-Sinoussi F, Montagutelli X. Animal models are essential to biological research: issues and perspectives. Future. Sci. OA. 1(4): FSO63 (2015).
- Gretebeck LM, Subbarao K. Animal models for SARS and MERS coronaviruses. Curr. Opin. Virol. 13: 123-129 (2015).
- Cockrell AS, Peck KM, Yount BL, et al. Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J. Virol. 88(9): 5195-5199 (2014)
- Nagata N, Iwata N, Hasegawa H, et al. Participation of both host and virus factors in induction of severe acute respiratory syndrome (SARS) in F344 rats infected with SARS coronavirus. J. Virol. 81(4): 1848-1857 (2007).
- Rahman, A. Atanu S. Risk Factors for Fatal Middle East Respiratory Syndrome Coronavirus Infections in Saudi Arabia: Analysis of the WHO Line List, 2013–2018. Am. J. Public. Health. 109(9): 1288-1293 (2019).
- Garcia-Cremades M, Solans BP, Hughes E, et al. Optimizing Hydroxychloroquine Dosing for Patients With COVID-19: An Integrative Modeling Approach for Effective Drug Repurposing. Clin. Pharmacol. Ther. 108(2): 253-263 (2020)
- Roden DM, Harrington RA, Poppas A, Russo AM. Considerations for Drug Interactions on QTc in Exploratory COVID-19 Treatment. Circulation. 141(24): 906-907 (2020).
- Bishara D, Kalafatis C, and Taylor D. Emerging and experimental treatments for COVID-19 and drug interactions with psychotropic agents. Ther. Adv. Psychopharmacol. 10: 2045125320935306 (2020).
- Bornstein SR, Rubino F, Khunti K, et al. Practical recommendations for the management of diabetes in patients with COVID-19. Lancet. Diabetes. Endocrinol. 8(6):546-550 (2020).
- Izmailova ES, Wagner JA, Perakslis ED. Wearable devices in clinical trials: hype and hypothesis. Clin. Pharmacol. Ther. 104(1):42-52 (2018).
- Nießl A, Beyersmann J, Loos A. Multistate modeling of clinical hold in randomized clinical trials. Pharm. Stat. 19(3): 262-275 (2020).







