
Responding to COVID-19: Recent Advances and Challenges in Diagnosis
*Corresponding Author(s):
Jie LiangXijing Hospital Of Digestive Diseases, 129 Chang-le West Road, Xi'an, Shaanxi Province, 710032, China
Tel:86-13488260468,
Email:liangjie@fmmu.edu.cn
Kaichun Wu
Xijing Hospital Of Digestive Diseases, 129 Chang-le West Road, Xi'an, Shaanxi Province, 710032, China
Tel:86-13709218743,
Email:kaicwu@fmmu.edu.cn
# Equal Contribution
Abstract
The pandemic of COVID-19 is a tremendous threat to global health. Clinical assessment, diagnostic testing, and necessary quarantine remain key measures to control the global pandemic. Currently, laboratory-based real-time RT-PCR assays for detecting SARS-CoV-2 are the cornerstone for COVID-19 diagnosis. There are several drawbacks to PCR testing, hence diagnosis of asymptomatic patients remains problematic. Therefore, several novel diagnostic strategies including serologic immunoassays, combined use of PCR and antibody testing, and point-of care molecular tests are rapidly emerging. The purpose of this review is to summarize the recent findings on the utility and limitations of current array of tests for SARS-CoV-2, highlighting the value of antibody test and fast diagnostic techniques. Also, the diagnostic values of rectal swabs and saliva are discussed.
Keywords
Covid-19; Immunoassays; Antibody
INTRODUCTION
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been declared a pandemic by the World Health Organization (WHO) and has created a tremendous global health crisis [1]. The number of COVID-19 cases is still escalating world-wide, as of May 4, 2020, there have been more than 3 435 894 laboratory-confirmed cases and more than 239 604 deaths reported in over 200 countries [2] .Healthcare workers from all over the world, including clinicians, nurses, laboratory technicians and coordination staff members are in the forefront of fighting against this public health crisis.
Although fever and cough were the most common symptoms, extra-pulmonary manifestations especially digestive symptoms could be the major complaint in a number of COVID-19 patients were noticed [3]. In the study by Wang et al. 14 cases out of 138 (10.1%) hospitalized COVID-19 patients had initial digestive symptoms of diarrhea and nausea, then fever and dyspnea [4]. And, a set of 6 cases from a cohort of 204 patients with COVID-19 were found to have only digestive symptoms, without respiratory symptoms [5]. Asymptomatic patients and patients with other atypical symptoms such as loss of sense of smell or taste also have been reported [6,7]. These findings have increased the uncertainty of the diagnostic work-up and raised concerns among clinicians. Moreover, given the lack of effective vaccines or treatments for COVID-19 until now, early identification of persons infected with SARS-CoV-2 virus followed by proper quarantine are the essential means to control the global pandemic.
Currently, in addition to symptomatic evaluation, laboratory confirmation of a COVID-19 case by real-time reverse transcription polymerase chain reaction (RT-PCR) remains the gold standard for the etiologic diagnosis of SARS-CoV-2 infection [8,9]. But several drawbacks of PCR should not be ignored as there can be false negative results. Serologic tests, or antibody tests are less complex than PCR tests and useful for confirming COVID-19 infection [10]. IgM may be a useful marker of more recent infection and IgG a reliable marker of past infection [11]. Combination of RNA and antibody testing significantly improves the diagnosis of SARS-CoV-2 infection with a specificity of as high as 99% [12]. With the nature of low-complexity, ambulatory and less time-consuming (giving results within one hour), the point-of-care (POC) molecular diagnostic test is a novel rapid diagnostic method. This rapid testing method has a potential to expand testing volumes across the world and could be useful in settings where clinical decisions require rapid results [13]. As viral shedding from the digestive system might be longer-lasting, there was a suggestion of the rectal swab assay included in the criteria for discharge or cease of quarantine of COVID-19 patients [14,15]. SARS-CoV-2 has been detected inself-collected saliva from most (84.6%-100%) infected patients even in those with negative nasopharyngeal aspirate PCR results [16-19]. Analyzing saliva samples may provide a promising non-invasive method for the diagnosis of COVID-19, especially useful in asymptomatic and mildly symptomatic patients, or for determine the appropriate period of quarantine for patients isolated at home.
The purpose of this review article is to summarize the update of diagnostic testing for SARS-CoV-2, highlighting the value of antibody test and fast diagnostic techniques, as the choice of a proper diagnostic testing plays a pivotal role in control of the pandemic of COVID-19.
DATA SOURCE AND LITERATURE SEARCH
The databases of PubMed, Web of Science, and Google Scholar were comprehensively searched for articles published from December 1st, 2019 to April 28th, 2020 with the following key words: “coronavirus disease 2019”, “COVID-19”, “severe acute respiratory syndrome corona virus 2”, “SARS-CoV-2”, “2019 novel coronavirus” or “2019-nCoV”; “Polymerase Chain Reaction”, “PCR”, “Diagnosis”, “serology”, “antibody” or “immune*” alone and in combination. Moreover, considering the fact at early stage of the outbreak majority of COVID-19 patients were Chinese and several findings were reported in Chinese journals, the following major Chinese medical databases including CNKI, WANFANG DATA, Sino Med were also systematic searched. Additional publications were manually searched in the reference lists of the included literature.
REAL-TIME RT-PCR ASSAYS
In previous clinical practice, PCR has been routinely performed to detect etiologic viruses of acute respiratory infections such as severe acute respiratory syndrome (SARS), novel human coronavirus (hCoV-EMC) infections and novel avian influenza A(H7N9) [20-22]. Similarly, positive-sense, single-stranded RNA genome of SARS-CoV-2 harbors several molecular targets which can be used for PCR assays, consisting of structural protein encoding genes and genes required for viral replication [8,9,23,24]. The structural proteins including spike protein (SP), envelope protein (EP), trans membrane protein (MP), and nucleocapsid protein (NP). In addition to genes encoding the above structural proteins, viral replication-associated genes involve RNA-dependent RNA polymerase (RdRp), hemagglutin inesterasedimer (HE), and open reading frames ORF1a and ORF1b [9,23-25]. Figure 1 shows the schematic of the SARS-CoV-2 [13]. Currently, PCR has been recommended as the corner stone of detection for viral RNA by CDC of the United States and WHO [9,26]. And the major advantage of PCR assays is the capability of minimizing false-positive results due to contamination of amplification product, because amplification and analysis of the specimen are done simultaneously in a closed system [27,28]. Figure 1 Schematic of the SARS-CoV-2 [13].
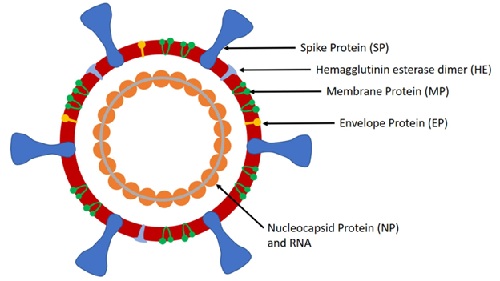
Figure 1: The schematic of the SARS-CoV-2.
The following three groups are recommended with testing priority: persons who had close contact with suspected or confirmed COVID-19 or had an affected area travel history, hospitalized patients with manifestations consistent with COVID-19, and other symptomatic persons who may suffer high risk of poor outcome [29]. Nasopharyngeal (NP) swab and/or oropharyngeal (OP) swab are commonly used testing specimen for acute respiratory infection [30,31]. But NP swab seems to be more preferred than OP, as the quality-control of specimen collection process can be easily achieved, namely reaching the right area to be tested in the nasal cavity and the procedure is safe to the operator. Moreover, the diagnostic performance has been reported to be better than that of OP. In the study by Wang et al., the SARS-CoV-2 60 RNA was identified in 63% of NP swabs, significantly higher than that in OP samples (32%) [32,33]. Specimens should be collected using a flocked swab, and the swab must be inserted deeply into the nasal cavity and “tears” are commonly elicited. Similarly, a gag reflex should be elicited during the OP swab collection process. After sample collection, specimens should be transferred timely for RNA extraction, then followed by real-time RT-PCR for target detection [34,35].
Currently, PCR has been mainly used for diagnosis of symptomatic patients. There are several drawbacks of this method that should not be ignored: 1) Viral RNA is usually obtained from a nasopharyngeal swab, but the viruses are predominantly a lower respiratory pathogen. It may not be present in sufficient quantity in the upper respiratory tract leading to false negative results. In this scenario, repeated testing or obtaining lower respiratory tract specimens such as sputum, endotracheal aspirates, and bronchoalveolar lavage (BAL) may be required [33,35] ; 2) The virus may be present in low titers in the incubation period, the negative predictive value of screening patients during incubation/asymptomatic phase is still unknown [35] ;3) The test requires expertise, consumables such as flocked swabs and/or transport media and sophisticated laboratory equipment which may be in short supply especially in the developing world;4) Whether a single time of negative result of upper respiratory tract swab can be adequate for ruling out COVID-19 remains unclear; 5) There remains an important gap in accurate determination of viral shedding in convalescence phase for de-isolation decisions; 6) Furthermore, the PCR is not useful in identifying patients who are post infection and may be immunized.
SEROLOGY TEST FOR SARS-COV-2
A humoral immune reaction with antibody production to pathogens is part of the normal host response. Serologic tests detect the antibodies (such as IgA, IgM, and IgG) to infection and is an indirect testing methods which has proven to be useful in the epidemiology of SARS and other virus outbreak [27,36,37]. SARS-CoV-2 has a number of antigenic sites, of which, spike protein (SP) and NP appear to be important ones for developing serological assays to detect COVID-19 [13,37]. ELISA and immune chromatographic methods have been commonly used to detect the antibodies (IgM and IgG) to these antigen sites and several novel methods such as automated chemiluminescent immunoassay (CLIA), manual ELISA, and rapid lateral flow immunoassay (LFIA) are rapidly emerging [13,27]. In early April, the United States (US) Food and Drug Administration (FDA) has given an emergency use authorization to serologic test. Also, the National Institutes of Health has launched a study to investigate the level of antibodies aiming to gather data for epidemiological models.
Serologic tests or antibody tests may be less complex than PCR tests and potentially useful for diagnosis in certain situations [10]. Currently, the main role of antibody testing lies in confirming COVID-19 infection. Given antibody production take days to weeks to be stably detectable, there is a concern that serology detection is not likely to be useful in the early phase of COVID-19 disease such as incubation or asymptomatic period. Regarding the time profile of antibody detection in patients with COVID-19, seroconversion was found to occur after 7 days in 50% of patients and 14 days in all [38]. Similarly, Xiao et al. analyzed the features of IgM and IgG from 34 COVID-19 patients in Wuhan using chemiluminescent immunoassay and found IgM last more than a month and IgG significantly longer. But only 2 patients in this study had their IgM and IgG tested in the first week after symptom onset, while the rest patients in this set were tested after 2 weeks from symptoms onset [10]. In another study including 214 patients with COVID-19, ELISA tests for IgM and IgG antibodies to recombinant nucleocapsid (rN) and spike (rS) proteins were conducted and the positive rates for rN based IgM and/or IgG was 80.4% and for rS 82.2%,respectively. Similarly, the positive rate for IgM begun to drop from day 35 after symptom onset and IgG persisted longer. Subsequently, the authors concluded that IgM may be a useful marker of more recent infection and IgG may be a reliable marker of past infection [11]. The combination of PCR and antibody testing described later may solve this problem.
Also, the following concerns deserve more efforts and evidence to clarify in order to define the utility of serologic test: 1) whether every patients who hasSARS-CoV-2 infection actually develops antibodies, whether the antibodies detected has a protect effect against secondary infections, and if so, how long the antibodies linger in the body, the answers to these questions remain unclear; 2) Potential cross-reactivity of antibody to non-SARS-CoV-2 coronavirus is also a problem and consequently, positive findings may be the result of past or present infection with other viruses; 3) IgM responses are often non-specific, while specific IgG antibody takes weeks to develop. Also, cross-reactivity to other previous coronavirus can result in a positive finding of IgG, even in patients with previously asymptomatic infection. In the future, serologic tests, when widely available and the concerns above clarified, will play an important role in epidemiologic studies, ongoing surveillance, vaccine development of COVID-19, and risk assessment of health care workers by determining the immune status.
COMBINED PCR AND ANTIBODY TESTING
Antibody testing has emerged as the second testing strategy adapted to aid the diagnosis of COVID-19. The combination of PCR and antibody testing can be the third approach.
In a study involving 173 COVID-19 patients, the presence of total antibodies (IgM and IgG) was found<40% in the first week after symptoms onset and rapidly increased to 100%. In contrast, the positive rate of RNA decreased from 66.7% before day 7 to 45.5% between days 15-39. Thus, the authors concluded combining RNA and antibody testing significantly improved the virus diagnosis. And the specificity was estimated at over 99% by testing serum from healthy individuals obtained before the outbreak of SARS-CoV-2 [12]. Further, the value of combining antibody and PCR testing was confirmed in another study, which stated that the detection rate increased to 98.6% for combined testing compared to 51.9% for a single PCR [39].
RAPID DIAGNOSTICS: POINT-OF-CARE (POC) MOLECULAR TEST
Point-of-care (POC) molecular diagnostics test is a novel rapid diagnostic method based on real-time RT-PCR or antibody test [13]. Low-complexity, ambulatory and less time-consuming (giving results within one hour) are the major strengths of this method. A variety of samples such as nasopharyngeal swab, nasal wash, or aspirate specimens can be used for detection [13,40]. As a result, POC diagnostics might be useful to expand testing scales across the world, especially in developing countries, and low-resource settings where clinical decisions require rapid results. To date, cartridge-based tests on platforms including Abbott ID NOW (Abbott Laboratories), BioFire Film Array (bio Merieux), cobas Liat (Roche Diagnostics), and Gene Xpert (Cepheid) have been approved for emergency use by US FDA [35,40]. Apart from SARS-CoV-2, a POC test developed by Bosch, Germany and Randox Laboratories, UK claimed able to simultaneously detect nine other respiratory virus like influenza A and B [41]. Another LFIA based POC test developed by Bio Medomics, USA requires a minimal sample volume (20ul of finger-picked blood and 10ul of serum/plasma) and can detect the IgM and IgG antibodies in 10 minutes. More importantly, neither trained personnel or any professional instrument is necessary to perform the test, thus it theoretically can be used at any place [42]. However, the testing throughput may hamper the value of POC in screening patients and in large-scale use.
CURRENT PRACTICE IN OUR CENTER
This section summarizes our experience in Xijing Hospital of digestive disease. The antibody assay used in our center is a product of Beijing Wantai Biological Pharmacy, which has been approved in early March by National Medical Products Administration of China and received a European Conformity (CE) certificate. The product covers 28 provinces with over 200,000 tests done in China, and 23countries with 35,000 tests done over the world. This is a CLIA based on double-antigen sandwich principle for specific antibody capture in the serum or plasma. The antigens used in this system are recombinant proteins containing the receptor-binding domain (RBD) of the SP ofSARS-CoV-2. Total antibodies are detected by applying 2 RBD proteins as the immobilized and HRP-conjugated antigen. The IgM μ chain capture method (IgM-ELISA) is used to detect the IgM antibodies. The assay takes 29 minutes to give the first result and the speed of detection can reach 200 tubes per hour if using automatic device of Caris200, Huawei Medical LLC, Beijing, China. The sensitivity and specificity of the kit is 94.8% and 99.7% respectively according to a previous study of 386 patients with confirmed COVID-19 and 1859 healthy controls [43].
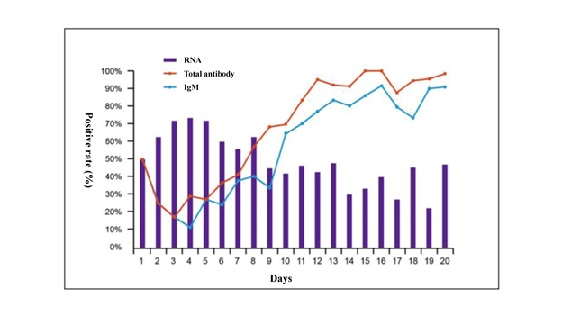
Figure 2: Positive rate of RNA detected by PCR using oropharyngeal and/or nasal swabs, total antibodies andIgM antibodies by days from onset of symptoms in patients with COVID-19 (Data onfile of Beijing Wantai Biological Pharmacy, submitted to National Medical Products Administration ofChina). N=380
The IgM positive rate (not quantification) curve exactly tracks the total antibody curve in the initial 20 days. Another study reported that the positive rate of IgG is about 19% in the first 7 days since symptom onset and rise to 80% by 30 days [12]. We assume the COVID-19 patients were not previously exposed to SARS-CoV-2, as this virus is new to humans and one possible explanation for the positive rate of 19% in the study by Zhao [12] and up to 50% in Figure 2 within seven days after symptom onset could be the cross-reactivity to other coronavirus although the assay claimed a 99% specificity with over 1800 healthy subjects as controls. The positive rate in Figure 2 falls to 50% in antibody response from first day of symptom onset to day3 to 4 post-on set may not mean anything as there were too few chances to have patients’ blood taken for antibody analysis at day 1 of symptom onset.
Of concern, 40% of patients had detectable RNA in swabs 20 days post-onset. We do not know how this relates to the risk of them transmitting infection but, if verified, this finding has implications with regards to the duration of isolation of affected patients. In practice, a study using the Wantai kit demonstrates a detection rate of 30%-40% for patients with COVID-19 within the initial 7 days, 70% at 8-10 days, and 100% through 12 days. In contrast to the antibody detection, the PCR assay in the same cohort of COVID-19 patients showed 60%-70% of positive rate within the initial 7 days and dropped to 40%-50% since then [12].
Based on the data provided by Beijing Wantai Biological Pharmacy, also submitted to National Medical Products Administration of China, the antibody status in confirmed COVID-19 patients with negative RNA results at different time points since onset of symptom are shown in Table 1.
|
Days of symptom onset |
RNA negative |
Antibody positive in patients with negative RNA result |
|
≤ 3 days |
7 |
2 |
|
4-7 days |
28 |
15 |
|
8-14 days |
57 |
56 |
|
≥ 15 days |
30 |
30 |
Table 1: Role of antibody testing in PCR negative patients.
PCR: Short for real-time reverse transcription polymerase chain reaction (RT-PCR).
In the first eight days post symptom onset, antibodies added an approximately 50% diagnostic gain in PCR negative patients. After eight days, antibodies were detected in nearly 100% of PCR negative patients. Thus, the antibody assay may be a complement to the PCR assay to identify patients with false negative results at early phase of disease. Currently, the approved indications of this antibody assay kit in China are:1) Additional testing of suspect patients with negative nucleic acid test of SARS-CoV-2; 2) Antibody tittering for patients recovered from COVID-19 [43].
We have started the SARS-CoV-2 antibody assay in my hospital since late March, mainly for screening for COVID-19 in every patient prior to his admission to hospital. We also performed this assay to screen patients before elective endoscopy. It has become a useful complement to the PCR assay and reduced the heavy burden on the clinical lab doing PCR testing by doing 400-500 tests of SARS-CoV-2 antibody assay every day.
OTHER CONSIDERATIONS: THE DIAGNOSTIC VALUE OF RECTAL SWABS AND SALIVA
Although, the majority of Covid-19 patients typically present with respiratory symptoms and signs, several suffering primarily with digestive symptoms including diarrhea, decline of appetite, nausea/vomiting and abdominal pain have been identified [25,44]. Considering the following facts: 1) the receptor of SARS-CoV-2, angiotens in converting enzyme 2 (ACE2), has been found highly expressed in gastrointestinal (GI) epithelial cells, and 2) the stool specimens of infected patients were determined harboring SARS-CoV-2 live viral RNA in patients with no GI symptoms, there is a concern about potential oral-fecal transmission of SARS-CoV-2 [26,33,45]. To date, several studies has found some patients can remain positive for SARS-CoV-2 in stool after their respiratory samples were negative [3,46], the authors therefore concluded viral shedding from the digestive system might be longer-lasting and emphasized the health care workers need to bear in mind that the stool might be infectious [14,15]. Similarly, by analyzing seven cases of COVID-19 who were readmitted to hospital because of positive RT-PCR after discharge, Zhang et al. found six patients had positive rectal swabs but negative upper respiratory tract swabs and suggested rectal swab assay be included in the criteria for discharge or cease of quarantine [47]. However, these findings are preliminary and further research is necessary.
Interestingly, the saliva has been found to be another alternative potential for detection of virus, and SARS-CoV-2 has been detected inself-collected saliva from most (84.6%-100%) infected patients even in those with negative nasopharyngeal aspirate PCR results [16-19]. Moreover, given the nature of non-invasive, easy collection, and less exposure of health provider, saliva for diagnostics is more acceptable for patients and secured for the staff who performed specimen collection and tests. Consequently, saliva may be a promising noninvasive specimen for the diagnosis of COVID-19, especially ideal for situations in which nasopharyngeal swabs collection are contraindicated. Further, to determine the role of salivary test in asymptomatic and mildly symptomatic patients is remarkably important. And if POC based on salivary test available, analyzing serial saliva samples which can be self-collected easily may be an attractive method to determine the appropriate period of quarantine for patients isolated at home.
CONCLUSION
Diagnostic testing for COVID-19 plays a central role in controlling the global pandemic. Laboratory-based real-time RT-PCR assays for detecting SARS-CoV-2 are the current main stay for COVID-19 diagnosis, and several novel diagnostic strategies including serologic immunoassays, combined use of PCR and antibody testing and POC molecular tests are rapidly emerging. However, early diagnosis of asymptomatic patients or patients in incubation phase remains a challenge. With regards to future directions the diagnostic values of rectal swabs and saliva deserve more investigation.
REFERENCES
- Padoan A, Cosma C, Sciacovelli L, Faggian D, Plebani M (2020) Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med.
- https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ .
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, et al. (2020) Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med.
- Wang D, Hu B, Hu C, Zhu F, Liu X, et al. (2020) Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA.
- Pan L, Mu M, Yang P, Sun Y, Wang R, et al. (2020) Clinical Characteristics of COVID-19 Patients With Digestive Symptoms in Hubei, China: A Descriptive, Cross-Sectional, Multicenter Study. Am J Gastroenterol 115: 766-773.
- Pan X, Chen D, Xia Y, Wu X, Li T, et al. (2020) Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect Dis 20: 410-411.
- Lechien JR, Chiesa Estomba CM, De Siati DR, Horoi M, Le Bon SD, et al. (2020) Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): A multicenter European study. Eur Arch Otorhinolaryngol.
- Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, et al. (2020) Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin Chem 66: 549-555.
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, et al. (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill.
- Xiao AT, Gao C, Zhang S (2020) Profile of Specific Antibodies to SARS-CoV-2: The First Report. J Infect.
- Liu W, Liu L, Kou G, Zheng Y, Ding Y, et al. (2020) Evaluation of Nucleocapsid and Spike Protein-based ELISAs for detecting antibodies against SARS-CoV-2. J Clin Microbiol.
- Zhao J, Yuan Q, Wang H, Liu W, Liao X, et al. (2020) Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis.
- Vashist SK (2020) In Vitro Diagnostic Assays for COVID-19: Recent Advances and Emerging Trends. Diagnostics (Basel) 10: E202.
- Xu Y, Li X, Zhu B, Liang H, Fang C, et al. (2020) Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med 26: 502-505.
- Hindson J (2020) COVID-19: Faecal-oral transmission? Nat Rev Gastroenterol Hepatol 17: 259.
- To KK, Tsang O, Chik Yan Yip C, Chan KH, Wu TC, et al. (2020) Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis.
- Azzi L, Carcano G, Gianfagna F, Grossi P, Gasperina DD, et al. (2020) Saliva is a reliable tool to detect SARS-CoV-2. J Infect.
- To KK, Tsang OT, Leung WS, Tam AR, Wu TC, et al. (2020) Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect Dis.
- Williams E, Bond K, Zhang B, Putland M, Williamson DA (2020) Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol.
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, et al. (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348: 1967-1976.
- Corman VM, Müller MA, Costabel U, Timm J, Binger T, et al. (2012) Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill 17.
- Corman VM, Eickmann M, Landt O, Bleicker T, Brünink S, et al. (2013) Specific detection by real-time reverse-transcription PCR assays of a novel avian influenza A(H7N9) strain associated with human spillover infections in China. Euro Surveill 18: 20461.
- Cui J, Li F, Shi ZL (2019) Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17: 181-192.
- Lu R, Zhao X, Li J, Niu P, Yang B, et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 395: 565-574.
- Chan JF, Yip CC, To KK, Tang TH7, Wong SC, et al. (2020) Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J Clin Microbiol 58: e00310-20.
- Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, et al. (2020) First Case of 2019 Novel Coronavirus in the United States. N Engl J Med 382: 929-936.
- Tang YW, Schmitz JE, Persing DH, Stratton CW (2020) The Laboratory Diagnosis of COVID-19 Infection: Current Issues and Challenges. J Clin Microbiol.
- Loeffelholz MJ, Tang YW (2020) Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg Microbes Infect 9: 747-756.
- Chen C, Zhang XR, Ju ZY, He WF (2020) [Advances in the research of cytokine storm mechanism induced by Corona Virus Disease 2019 and the corresponding immunotherapies]. Zhonghua Shao Shang Za Zhi 36: E005.
- Zou L, Ruan F, Huang M, Liang L, Huang H, et al. (2020) SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med 382: 1177-1179.
- Chan PK, To WK, Ng KC, Lam RK, Ng TK, et al. (2004) Laboratory diagnosis of SARS. Emerg Infect Dis 10: 825-831.
- Kim C, Ahmed JA, Eidex RB, Nyoka R, Waiboci LW, et al. (2011) Comparison of nasopharyngeal and oropharyngeal swabs for the diagnosis of eight respiratory viruses by real-time reverse transcription-PCR assays. PLoS One 6: e21610.
- Wang W, Xu Y, Gao R, Lu R, Han K, et al. (2020) Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA.
- Druce J, Garcia K, Tran T, Papadakis G, Birch C (2012) Evaluation of swabs, transport media, and specimen transport conditions for optimal detection of viruses by PCR. J Clin Microbiol 50: 1064-1065.
- Cheng MP, Papenburg J, Desjardins M, Kanjilal S, Quach C, et al. (2020) Diagnostic Testing for Severe Acute Respiratory Syndrome-Related Coronavirus-2: A Narrative Review. Ann Intern Med.
- Chen X, Zhou B, Li M, Liang X, Wang H, et al. (2004) Serology of severe acute respiratory syndrome: Implications for surveillance and outcome. J Infect Dis 189: 1158-1163.
- Chan CM, Tse H, Wong SS, Woo PC, Lau SK, et al. (2009) Examination of seroprevalence of coronavirus HKU1 infection with S protein-based ELISA and neutralization assay against viral spike pseudotyped virus. J Clin Virol 45: 54-60.
- Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, et al. (2020) Virological assessment of hospitalized patients with COVID-2019. Nature.
- Guo L, Ren L, Yang S, Xiao M, Chang, et al. (2020) Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin Infect Dis.
- Hogan CA, Caya C, Papenburg J (2018) Rapid and simple molecular tests for the detection of respiratory syncytial virus: A review. Expert Rev Mol Diagn18: 617-629.
- Chenoweth AM, Wines BD, Anania JC, Mark Hogarth P (2020) Harnessing the immune system via FcgammaR function in immune therapy: A pathway to next-gen mAbs. Immunol Cell Biol 98: 287-304.
- Spranger S, Sznol M, Weiner GJ, Wiggington JM, Weber JS, et al. (2020) Is antibody-dependent enhancement playing a role in COVID-19 pathogenesis? J Immunother Cancer150: w20249.
- Cheng J, Chen Z, Sun K, Pan W, Zhan Z, et al. (2020) Is COVID-19 receiving ADE from other coronaviruses? J Med Virol 22: 72-73.
- Wu Y, Guo C, Tang L, Hong Z, Zhou J, et al. (2020) Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 5: 434-435.
- Xiao F, Tang M, Zheng X, Liu Y, Li X, et al. (2020) Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 158: 1831-1833.e3.
- Yeo C, Kaushal S, Yeo D (2020) Enteric involvement of coronaviruses: Is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol 5: 335-337.
- Zhang B, Liu S, Dong Y, Zhang L, Zhong Q, et al. (2020) Positive rectal swabs in young patients recovered from coronavirus disease 2019 (COVID-19). J Infect.
Citation: Su S (2020) Responding to COVID-19: Recent Advances and Challenges in Diagnosis. J Clin Immunol Immunother 6: 024.
Copyright: © 2020 Song Su, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

