Proteomic Analysis of Human Milk Reveals Nutritional and Immune Benefits in the Colostrum from Mothers with COVID-19
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Milk Sample Preparation for Proteomic Analysis
2.3. Liquid Chromatography–Tandem Mass Spectrometry
2.4. MS Settings for Proteomic Analysis
2.5. MS Settings for Glycoproteomic Analysis
2.6. Data Processing
2.7. Statistical Analysis
2.8. Availability of Data and Materials
3. Results
3.1. Alteration of Proteome Profiles in Milk from COVID Patients
3.2. Upregulation of Immune-Related Proteins Suggest the Adaptive Immune Benefit of Colostrum from COVID-19 Patients
3.3. The Increased Microheterogeneity of Overall Protein Glycosylation and the Glycosylation Levels of Immune-Related Proteins in the COVID-19 Colostrum Proteome
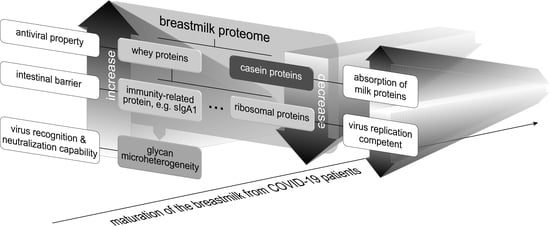
3.4. COVID-19 Colostrum’s Immune-Related Proteome Profile Changes Fade with Milk Maturation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, N.; van Unen, V.; Abdelaal, T.; Guo, N.; Kasatskaya, S.A.; Ladell, K.; McLaren, J.E.; Egorov, E.S.; Izraelson, M.; Chuva de Sousa Lopes, S.M.; et al. Memory CD4(+) T cells are generated in the human fetal intestine. Nat. Immunol. 2019, 20, 301–312. [Google Scholar] [CrossRef] [PubMed]
- WHO. COVID-19 Clinical Management: Living Guidance, 25 January 2021; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Wang, Y.; Chen, L.; Wu, T.; Shi, H.; Li, Q.; Jiang, H.; Zheng, D.; Wang, X.; Wei, Y.; Zhao, Y.; et al. Impact of Covid-19 in pregnancy on mother’s psychological status and infant’s neurobehavioral development: A longitudinal cohort study in China. BMC Med. 2020, 18, 347. [Google Scholar] [CrossRef] [PubMed]
- Lönnerdal, B. Nutritional and physiologic significance of human milk proteins. Am. J. Clin. Nutr. 2003, 77, 1537s–1543s. [Google Scholar] [CrossRef] [PubMed]
- Tromp, I.; Kiefte-de Jong, J.; Raat, H.; Jaddoe, V.; Franco, O.; Hofman, A.; de Jongste, J.; Moll, H. Breastfeeding and the risk of respiratory tract infections after infancy: The Generation R Study. PLoS ONE 2017, 12, e0172763. [Google Scholar] [CrossRef] [Green Version]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Woof, J.M.; Russell, M.W. Structure and function relationships in IgA. Mucosal Immunol. 2011, 4, 590–597. [Google Scholar] [CrossRef] [Green Version]
- Shehata, L.; Wieland-Alter, W.F.; Maurer, D.P.; Chen, E.; Connor, R.I.; Wright, P.F.; Walker, L.M. Systematic comparison of respiratory syncytial virus-induced memory B cell responses in two anatomical compartments. Nat. Commun. 2019, 10, 1126. [Google Scholar] [CrossRef] [Green Version]
- Chantry, C.J.; Howard, C.R.; Auinger, P. Full breastfeeding duration and associated decrease in respiratory tract infection in US children. Pediatrics 2006, 117, 425–432. [Google Scholar] [CrossRef]
- Georgi, G.; Bartke, N.; Wiens, F.; Stahl, B. Functional glycans and glycoconjugates in human milk. Am. J. Clin. Nutr. 2013, 98, 578S–585S. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, H.; Yamashita, Y. Role of N-glycosylation in cell surface expression and protection against proteolysis of the intestinal anion exchanger SLC26A3. Am. J. Physiol. Cell Physiol. 2012, 302, C781–C795. [Google Scholar] [CrossRef]
- Liu, B.; Newburg, D.S. Human milk glycoproteins protect infants against human pathogens. Breastfeed. Med. 2013, 8, 354–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, D.L. The Role of Human Milk Immunomodulators in Protecting Against Viral Bronchiolitis and Development of Chronic Wheezing Illness. Children 2015, 2, 289–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, D.S.; Mann, M.; Melby, J.A.; Larson, E.J.; Zhu, Y.; Brasier, A.R.; Jin, S.; Ge, Y. Structural O-Glycoform Heterogeneity of the SARS-CoV-2 Spike Protein Receptor-Binding Domain Revealed by Top-Down Mass Spectrometry. J. Am. Chem. Soc. 2021, 143, 12014–12024. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lin, Y.H.; Dingess, K.A.; Mank, M.; Stahl, B.; Heck, A.J.R. Quantitative Longitudinal Inventory of the N-Glycoproteome of Human Milk from a Single Donor Reveals the Highly Variable Repertoire and Dynamic Site-Specific Changes. J. Proteome Res. 2020, 19, 1941–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucl. Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahrne, E.; Molzahn, L.; Glatter, T.; Schmidt, A. Critical assessment of proteome-wide label-free absolute abundance estimation strategies. Proteomics 2013, 13, 2567–2578. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Mlecnik, B.; Galon, J.; Bindea, G. Comprehensive functional analysis of large lists of genes and proteins. J. Proteom. 2018, 171, 2–10. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucl. Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Guo, J.; Wang, C.; Luo, F.; Yu, X.; Zhang, W.; Li, J.; Zhao, D.; Xu, D.; Gong, Q.; et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: A retrospective review of medical records. Lancet 2020, 395, 809–815. [Google Scholar] [CrossRef] [Green Version]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. N. Am. 2013, 60, 49–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lönnerdal, B.; Erdmann, P.; Thakkar, S.K.; Sauser, J.; Destaillats, F. Longitudinal evolution of true protein, amino acids and bioactive proteins in breast milk: A developmental perspective. J. Nutr. Biochem. 2017, 41, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Weber, D.; Xu, W.; Durbin-Johnson, B.P.; Phinney, B.S.; Lönnerdal, B. Absolute Quantification of Human Milk Caseins and the Whey/Casein Ratio during the First Year of Lactation. J. Proteome Res. 2017, 16, 4113–4121. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Dingess, K.A. The Functional Power of the Human Milk Proteome. Nutrients 2019, 11, 1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoen, E.N.-T.; Cremer, T.; Gallo, R.C.; Margolis, L.B. Extracellular vesicles and viruses: Are they close relatives? Proc. Natl. Acad. Sci. USA 2016, 113, 9155–9161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, F.E.; Braathen, R.; Brandtzaeg, P. Role of J chain in secretory immunoglobulin formation. Scand. J. Immunol. 2000, 52, 240–248. [Google Scholar] [CrossRef]
- Pancera, M.; Shahzad-ul-Hussan, S.; Doria-Rose, N.A.; McLellan, J.S.; Bailer, R.T.; Dai, K.; Loesgen, S.; Louder, M.K.; Staupe, R.P.; Yang, Y.; et al. Structural basis for diverse N-glycan recognition by HIV-1–neutralizing V1–V2–directed antibody PG16. Nat. Struct. Mol. Biol. 2013, 20, 804. [Google Scholar] [CrossRef] [Green Version]
- Fouda, G.G.; Jaeger, F.H.; Amos, J.D.; Ho, C.; Kunz, E.L.; Anasti, K.; Stamper, L.W.; Liebl, B.E.; Barbas, K.H.; Ohashi, T. Tenascin-C is an innate broad-spectrum, HIV-1–neutralizing protein in breast milk. Proc. Natl. Acad. Sci. USA 2013, 110, 18220–18225. [Google Scholar] [CrossRef] [Green Version]
- Dierick, M.; Vanrompay, D.; Devriendt, B.; Cox, E. Lactoferrin, a versatile natural antimicrobial glycoprotein that modulates the host’s innate immunity. Biochem. Cell Biol. 2021, 99, 61–65. [Google Scholar] [CrossRef]
- Wockel, A.; Abou-Dakn, M.; Beggel, A.; Arck, P. Inflammatory breast diseases during lactation: Health effects on the newborn-a literature review. Mediat. Inflamm. 2008, 2008, 298760. [Google Scholar] [CrossRef] [Green Version]
- Riskin, A.; Almog, M.; Peri, R.; Halasz, K.; Srugo, I.; Kessel, A. Changes in immunomodulatory constituents of human milk in response to active infection in the nursing infant. Pediatr. Res. 2012, 71, 220–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; de Waard, M.; Verheijen, H.; Boeren, S.; Hageman, J.A.; van Hooijdonk, T.; Vervoort, J.; van Goudoever, J.B.; Hettinga, K. Changes over lactation in breast milk serum proteins involved in the maturation of immune and digestive system of the infant. J. Proteom. 2016, 147, 40–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; McMahon, R.J.; Woo, J.G.; Davidson, B.S.; Morrow, A.L.; Zhang, Q. Temporal changes in milk proteomes reveal developing milk functions. J. Proteome Res. 2012, 11, 3897–3907. [Google Scholar] [CrossRef] [PubMed]
- Raheem, R.A.; Binns, C.W.; Chih, H.J. Protective effects of breastfeeding against acute respiratory tract infections and diarrhoea: Findings of a cohort study. J. Paediatr. Child Health 2017, 53, 271–276. [Google Scholar] [CrossRef]
- Groß, R.; Conzelmann, C.; Müller, J.A.; Stenger, S.; Steinhart, K.; Kirchhoff, F.; Münch, J. Detection of SARS-CoV-2 in human breastmilk. Lancet 2020, 395, 1757–1758. [Google Scholar] [CrossRef]
- Tam, P.C.K.; Ly, K.M.; Kernich, M.L.; Spurrier, N.; Lawrence, D.; Gordon, D.L.; Tucker, E.C. Detectable severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in human breast milk of a mildly symptomatic patient with coronavirus disease 2019 (COVID-19). Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 72, 128–130. [Google Scholar] [CrossRef]
- Costa, S.; Posteraro, B.; Marchetti, S.; Tamburrini, E.; Carducci, B.; Lanzone, A.; Valentini, P.; Buonsenso, D.; Sanguinetti, M.; Vento, G.; et al. Excretion of SARS-CoV-2 in human breast milk. Clin. Microbiol. Infect. 2020, 26, 1430–1432. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, W.; Su, H.; Li, S.; Shereen, M.A.; Lv, Z.; Niu, Z.; Li, D.; Liu, F.; Luo, Z.; et al. Breastfeeding Risk from Detectable Severe Acute Respiratory Syndrome Coronavirus 2 in Breastmilk. J. Infect. 2020, 81, 452–482. [Google Scholar] [CrossRef]
- Wang, L.; Shi, Y.; Xiao, T.; Fu, J.; Feng, X.; Mu, D.; Feng, Q.; Hei, M.; Hu, X.; Li, Z.; et al. Chinese expert consensus on the perinatal and neonatal management for the prevention and control of the 2019 novel coronavirus infection (First edition). Ann. Transl. Med. 2020, 8, 47. [Google Scholar] [CrossRef]
- Perrine, C.G.; Chiang, K.V.; Anstey, E.H.; Grossniklaus, D.A.; Boundy, E.O.; Sauber-Schatz, E.K.; Nelson, J.M. Implementation of Hospital Practices Supportive of Breastfeeding in the Context of COVID-19—United States, 15 July–20 August 2020. MMWR. Morb. Mortal. Wkly. Rep. 2020, 69, 1767–1770. [Google Scholar] [CrossRef]
- Fujita, M.; Roth, E.; Lo, Y.J.; Hurst, C.; Vollner, J.; Kendell, A. In poor families, mothers’ milk is richer for daughters than sons: A test of Trivers-Willard hypothesis in agropastoral settlements in Northern Kenya. Am. J. Phys. Anthropol. 2012, 149, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Michaelsen, K.F.; Skafte, L.; Badsberg, J.H.; Jørgensen, M. Variation in macronutrients in human bank milk: Influencing factors and implications for human milk banking. J. Pediatr. Gastroenterol. Nutr. 1990, 11, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Shanes, E.D.; Mithal, L.B.; Otero, S.; Azad, H.A.; Miller, E.S.; Goldstein, J.A. Placental Pathology in COVID-19. Am. J. Clin. Pathol. 2020, 154, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.C.; Struve, M.; Raff, H. Effects of hypoxia on the development of intestinal enzymes in neonatal and juvenile rats. Exp. Biol. Med. 2003, 228, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Osorio, J.S. Gut health, stress, and immunity in neonatal dairy calves: The host side of host-pathogen interactions. J. Anim. Sci. Biotechnol. 2020, 11, 105. [Google Scholar] [CrossRef]
- Lönnerdal, B. Human milk proteins: Key components for the biological activity of human milk. Adv. Exp. Med. Biol. 2004, 554, 11–25. [Google Scholar]
- Lönnerdal, B. Bioactive Proteins in Human Milk: Health, Nutrition, and Implications for Infant Formulas. J. Pediatr. 2016, 173, S4–S9. [Google Scholar] [CrossRef] [Green Version]
- Dingess, K.A.; Gazi, I.; van den Toorn, H.W.P.; Mank, M.; Stahl, B.; Reiding, K.R.; Heck, A.J.R. Monitoring Human Milk β-Casein Phosphorylation and O-Glycosylation Over Lactation Reveals Distinct Differences between the Proteome and Endogenous Peptidome. Int. J. Mol. Sci. 2021, 22, 8140. [Google Scholar] [CrossRef]
- Fan, H.; Hong, B.; Luo, Y.; Peng, Q.; Wang, L.; Jin, X.; Chen, Y.; Hu, Y.; Shi, Y.; Li, T.; et al. The effect of whey protein on viral infection and replication of SARS-CoV-2 and pangolin coronavirus in vitro. Signal Transduct. Target Ther. 2020, 5, 275. [Google Scholar] [CrossRef]
- Brandtzaeg, P. The mucosal immune system and its integration with the mammary glands. J. Pediatr. 2010, 156 (Suppl. 2), S8–S15. [Google Scholar] [CrossRef]
- Goldman, A.S. Modulation of the gastrointestinal tract of infants by human milk. Interfaces and interactions. An evolutionary perspective. J. Nutr. 2000, 130 (Suppl. S2), 426s–431s. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.E.; Arend, F.M.; Doll, S.; Louiset, M.-L.; Winter, S.V.; Müller-Reif, J.B.; Torun, F.M.; Weigand, M.; Eichhorn, P.; Bruegel, M.; et al. High-resolution longitudinal serum proteome trajectories in COVID-19 reveal patients-specific seroconversion. BMJ MedRxiv 2021. [Google Scholar] [CrossRef]
- Van Keulen, B.J.; Romijn, M.; Bondt, A.; Dingess, K.A.; Kontopodi, E.; van der Straten, K.; den Boer, M.A.; Burger, J.A.; Poniman, M.; Bosch, B.J.; et al. Human Milk from Previously COVID-19-Infected Mothers: The Effect of Pasteurization on Specific Antibodies and Neutralization Capacity. Nutrients 2021, 13, 1645. [Google Scholar] [CrossRef] [PubMed]
- Pace, R.M.; Williams, J.E.; Järvinen, K.M.; Belfort, M.B.; Pace, C.D.W.; Lackey, K.A.; Gogel, A.C.; Nguyen-Contant, P.; Kanagaiah, P.; Fitzgerald, T.; et al. Characterization of SARS-CoV-2 RNA, Antibodies, and Neutralizing Capacity in Milk Produced by Women with COVID-19. mBio 2021, 12, e03192-20. [Google Scholar] [CrossRef]
- Hurley, W.L.; Theil, P.K. Perspectives on immunoglobulins in colostrum and milk. Nutrients 2011, 3, 442–474. [Google Scholar] [CrossRef]
- Hanson, L.A.; Korotkova, M. The role of breastfeeding in prevention of neonatal infection. Semin. Neonatol. SN 2002, 7, 275–281. [Google Scholar] [CrossRef]
- Mantis, N.J.; Rol, N.; Corthesy, B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Muco. Immunol. 2011, 4, 603–611. [Google Scholar] [CrossRef]
- Van de Perre, P. Transfer of antibody via mother’s milk. Vaccine 2003, 21, 3374–3376. [Google Scholar] [CrossRef]
- Newburg, D.S. Innate immunity and human milk. J. Nutr. 2005, 135, 1308–1312. [Google Scholar] [CrossRef] [Green Version]
- Fox, A.; Marino, J.; Amanat, F.; Krammer, F.; Hahn-Holbrook, J.; Zolla-Pazner, S.; Powell, R.L. Robust and Specific Secretory IgA Against SARS-CoV-2 Detected in Human Milk. iScience 2020, 23, 101735. [Google Scholar] [CrossRef]
- Narayanaswamy, V.; Pentecost, B.; Alfandari, D.; Chin, E.; Minor, K.; Kastrinakis, A.; Lieberman, T.; Arcaro, K.F.; Leftwich, H. Humoral and Cell-Mediated Immune Response in Colostrum from Women Diagnosed Positive for SARS-CoV-2. Breastfeed. Med. 2021, 16, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Lee, C. Regulatory Nucleotide Sequence Signals for Expression of the Genes Encoding Ribosomal Proteins. Front. Genet. 2020, 11, 501. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.R.; Scaiola, A.; Loughran, G.; Leibundgut, M.; Kratzel, A.; Meurs, R.; Dreos, R.; O’Connor, K.M.; McMillan, A.; Bode, J.W.; et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 2021, 372, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, D.; Wang, Y.; Su, W.; Liu, G.; Dong, W. The Importance of Glycans of Viral and Host Proteins in Enveloped Virus Infection. Front. Immunol. 2021, 12, 638573. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Grant, O.C.; Montgomery, D.; Ito, K.; Woods, R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020, 10, 14991. [Google Scholar] [CrossRef]
- Yang, Q.; Hughes, T.A.; Kelkar, A.; Yu, X.; Cheng, K.; Park, S.; Huang, W.C.; Lovell, J.F.; Neelamegham, S. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. Elife 2020, 9, e61552. [Google Scholar] [CrossRef]
- Wielgat, P.; Rogowski, K.; Godlewska, K.; Car, H. Coronaviruses: Is Sialic Acid a Gate to the Eye of Cytokine Storm? From the Entry to the Effects. Cells 2020, 9, 1963. [Google Scholar] [CrossRef]



| Characteristics | Case (n = 6) | Control (n = 10) | p-Value | |
|---|---|---|---|---|
| Maternal | age, median (IQR a) (years) | 30.00 (27.75, 30.00) | 33.50 (31.00, 34.75) | 0.08 |
| pre-pregnancy BMI b, median (IQR) (kg/m2) | 19.00 (18.23, 22.03) | 21.55 (20.35, 22.13) | 0.22 | |
| gestational weight gain, median (IQR) (kg) | 18.50 (15.75, 19.00) | 14.00 (12.25, 15.75) | 0.06 | |
| multiparous, n (%) | 3 (50) | 8 (80) | 0.30 | |
| undergraduate, n (%) | 3 (50) | 7 (70) | 0.61 | |
| postgraduate, n (%) | 3 (50) | 3 (30) | 0.61 | |
| Diseases during pregnancy | anemia, n (%) | 0 (0) | 0 (0) | NA |
| diabetes, n (%) | 1 (17) | 3 (30) | 1.00 | |
| hypertension, n (%) | 0 (0) | 0 (0) | NA | |
| pre-eclampsia, n (%) | 1 (17) | 0 (0) | 0.38 | |
| Infant | male, n (%) | 3 (50) | 6 (60) | 1.00 |
| gestational age on admission, median (IQR) (week) | 37.50 (37.00, 38.75) | 39.00 (38.25, 39.00) | 0.24 | |
| birth weight, median (IQR) (kg) | 2.99 (2.79, 3.14) | 3.24 (3.10, 3.62) | 0.07 | |
| birth length, median (IQR) (cm) | 49.50 (48.25, 50) | 49.00 (48.00,49.75) | 0.82 | |
| head circumference, median (IQR) (cm) | 34.50 (34.00, 35.00) | 35.00 (35.00, 35.25) | 0.08 | |
| Apgar 1-min, median (IQR) | 9 (9, 9) | 9 (9, 10) | 0.08 | |
| Apgar 5-min, median (IQR) | 10 (10, 10) | 10 (10, 10) | 0.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.; Tan, M.; Zhu, J.; Tian, Y.; Liu, H.; Luo, F.; Wang, J.; Huang, Y.; Zhang, Y.; Yang, Y.; et al. Proteomic Analysis of Human Milk Reveals Nutritional and Immune Benefits in the Colostrum from Mothers with COVID-19. Nutrients 2022, 14, 2513. https://doi.org/10.3390/nu14122513
Guo J, Tan M, Zhu J, Tian Y, Liu H, Luo F, Wang J, Huang Y, Zhang Y, Yang Y, et al. Proteomic Analysis of Human Milk Reveals Nutritional and Immune Benefits in the Colostrum from Mothers with COVID-19. Nutrients. 2022; 14(12):2513. https://doi.org/10.3390/nu14122513
Chicago/Turabian StyleGuo, Juanjuan, Minjie Tan, Jing Zhu, Ye Tian, Huanyu Liu, Fan Luo, Jianbin Wang, Yanyi Huang, Yuanzhen Zhang, Yuexin Yang, and et al. 2022. "Proteomic Analysis of Human Milk Reveals Nutritional and Immune Benefits in the Colostrum from Mothers with COVID-19" Nutrients 14, no. 12: 2513. https://doi.org/10.3390/nu14122513