Abstract
Background
We report here the case of two coworkers infected by the same SARS-CoV-2 strain, presenting two different immunological outcomes.
Case
One patient presented a strong IgG anti-receptor-binding domain immune response correlated with a low and rapidly decreasing titer of neutralizing antibodies. The other patient had a similar strong IgG anti-receptor-binding domain immune response but high neutralizing antibody titers.
Discussion and conclusion
Thus, host individual factors may be the main drivers of the immune response varying with age and clinical severity.
Similar content being viewed by others
Introduction
The emerging severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) in China at the end of 2019 has caused the current pandemic of coronavirus infectious disease (COVID)-19, which has infected every continent inhabited by virus specific immunologically-naïve humans [1].
The immune system induction following the SARS-CoV-2 infection is still not well known, although it is established that between 11 and 19 days after onset of symptoms most patients get a specific and neutralizing antibody (Ab) response [2]. The dynamic of IgM and IgG specific immune responses can vary along different factors, leading to various clinical severities of disease [3, 4]. Neutralizing Abs (nAbs) are of paramount importance for virus clearance, but their role in COVID-19 is not clearly established [5]. In most studies however, the specific antibody response is correlated with the emergence of nAbs [6, 7].
We report here the case of two co-workers, infected with the same SARS-CoV-2 strain, presenting two different clinical pictures and immunological outcomes. Interestingly, in one case the IgG response was not correlated with the detection of nAbs in our assay.
Case report
Patient 1 was a 26 years old female with no known risk factor. She presented on April 7, 2020 an isolated anosmia-agueusia. Three days later she felt a deep asthenia. She tested positive for SARS-CoV-2 by reverse transcriptase-polymerase chain reaction (RT-PCR) on April 12, 2020. She continued to experience a profound asthenia for 15 days, and completely healed except for the dysosmia, which was still partially present at day 100.
Patient 2 was a male, 51 with no risk factor besides age. He worked with patient 1 on April 8. He started to slightly cough on April 11, 2020. The following day, he felt tired, sub-febrile with an increasing cough. He consulted for a suspicion of COVID-19 at a hospital emergency department on April 12, 2020. At the initial examination, patient 2 had a polypnea at 32 respirations/min. The blood gas showed a hypoxemia with a PpO2 at 72 mmHg, PpCO2 42 mmHg, while a lymphopenia at 680 lymphocytes/mm3 was noted on the blood cell count. The chest computed-tomography scanner was normal, and the nasopharyngeal RT-PCR was positive for SARS-CoV-2. Patient 2 was discharged from the emergency room with a diagnosis of a mild form of COVID-19. He recovered at home within 15 days without major clinical complication, besides a month-long asthenia.
The SARS-CoV-2 strains from both patients were sequenced from naso-pharyngeal samples by MinION technology, following Artic protocol by PCR tiling [8]. Data were analyzed according to the bio-informatic protocol developed by the Artic consortium. Both patients were infected by the same strain, its sequence harboring 7 SNPs compared to the reference genome Wuhan/Hu-1/2019 (NCBI Nucleotide—NC_045512, GenBank—MN908947) and belonging to the G3b phylum [9], thus carrying the recently identified D614G mutation [10]. On August 1st and 2nd, 2020, the two sequences were deposited on the GISAID platform with accession ID EPI_ISL_505003 and EPI_ISL_506041 for patient 1 and 2 respectively.
The humoral immune response of both patients was followed serially for up to 100 days. An in-house enzyme-linked immuno-sorbent assay (ELISA) was developed for detecting IgG against SARS-CoV-2, adapted from the previous works of Florian Krammer team [11]. The ELISA detection was based on the receptor-binding domain (RBD) of the SARS-CoV-2 spike (S)-glycoprotein. ELISA results are presented as optical density (OD) ratio obtained by dividing the average OD of duplicate wells from that of the corresponding blank non-coated wells.
For each time point, the presence of nAbs was also sought by a seroneutralisation assay performed on Vero cells using the Institut Pasteur SARS-CoV-2 reference strain, in a BSL3 facility.
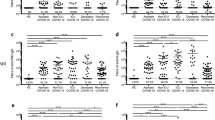
Both patients rapidly developed an IgG immune response against RBD as they were positive within 12 days, then marked a steep increase followed by a plateau and a slow decrease (Fig. 1). Patient 1 had a stronger IgG anti-RBD response while presenting a pauci-symptomatic infection. Patient 2 had also a robust anti-RBD response, while presenting mild clinical symptoms, that included blood desaturation as measured initially. Strikingly, patient 1 did only develop a very moderate neutralizing immune response with low nAb titers that turned negative by day 100, suggesting that virus clearance and the clinical recovery occurred independently of the nAb response.
Patients 1 and 2 IgG ELISA OD ratio against SARS-CoV-2 and seroneutralizing titers. a Green triangle, OD ratio RBD signal patient 1 (RBD P1); blue triangle, OD ratio RBD signal patient 2 (RBD P2). b Green sphere, seroneutralizing titers patient 1 (SN P1); blue sphere, seroneutralizing titers patient 2 (SN P2)
Discussion
The case presented underscores the role of unknown individual host factors or the potential other human coronavirus past infection in the serological response against SARS-CoV-2. As both patients were infected in a close time-line by the same virus, we can rule out the role of any virus effect on the immune response.
The protective role of antibodies and cellular immune response against SARS-CoV-2 is unknown, but antibodies are usually a reasonable correlate of immunity [2]. As RBD is the site of interaction of the S-glycoprotein with the angiotensin-converting enzyme (ACE)-2, which plays the role of virus receptor, a large number of neutralizing epitopes are located on RBD [12, 13]. In the vast majority of studies, the anti-RBD antibody levels correlate to serum viral neutralizing activity [6, 7].
Conclusion
Here, we demonstrate that RBD antibody levels and nAbs might not be correlated. The age, the clinical severity of the case [14], the sex [15], and several individual unknown host factors may influence the strength of the immune responses independently of the virus genotype.
Some other questions are still to be answered such as the long-term duration of the Ab response, and the correlate of protection. These data are crucial for evaluating the variability of the immunity in future vaccine studies.
References
Morens DM, Daszak P, Markel H, Taubenberger JK. Pandemic COVID-19 joins history’s pandemic legion. mBio. 2020;11:3.
Vabret N, Britton GJ, Gruber C, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–41.
Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–8.
To KKW, Tsang OTY, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–74.
Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–9.
Okba NMA, Muller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26:1478–88.
Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–9.
Quick J, Grubaugh ND, Pullan ST, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12:1261–76.
Gambaro F, Behillil S, Baidaliuk A, et al. Introductions and early spread of SARS-CoV-2 in France, 24 January to 23 March 2020. Euro Surveill. 2020;25:26.
Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–27.
Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26:1033–6.
Yuan M, Wu NC, Zhu X, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–3.
Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–9.
Wang X, Guo X, Xin Q, et al. Neutralizing antibody responses to severe acute respiratory syndrome coronavirus 2 in coronavirus disease 2019 inpatients and convalescent patients. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa721
Park MD. Sex differences in immune responses in COVID-19. Nature Rev Immunol. 2020;20:461.
Funding
This work was supported by the Délégation générale pour l’armement [Biomedef COVID5].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None of the authors has any conflict of interest relevant to the content of this work. There are no competing interests.
Rights and permissions
About this article
Cite this article
Billon-Denis, E., Ferrier-Rembert, A., Garnier, A. et al. Differential serological and neutralizing antibody dynamics after an infection by a single SARS-CoV-2 strain. Infection 49, 781–783 (2021). https://doi.org/10.1007/s15010-020-01556-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-020-01556-8