- *Corresponding Author:
- A. Bhardwaj
Department of Biotechnology & Life Sciences
Institute of Biomedical Education & Research
Mangalayatan University, Aligarh- 202145
Uttar Pradesh, India
E-mail: ankurphd995@gmail.com
| Date of Received | 10 August 2020 |
| Date of Revision | 15 December 2020 |
| Date of Acceptance | 20 February 2021 |
| Indian J Pharm Sci 2021;83(1):21-31 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Contagious human coronavirus belong to family Coronaviridae and infects human respiratory system causing the disease known as COVID-19 (WHO). To eradicate the severe acute respiratory syndrome coronavirus 2 pandemic, an effective vaccine should be developed. In the current study immunoinformatics procedures were employed to introduce a novel multi-epitope subunit vaccine. This multi-epitope vaccine can activate equally class I and II human leukocyte antigen and antibody mediated immune responses. Spike glycoprotein (PDB Id: 6VSB) of the coronavirus was selected and analysed using immune epitope database server for prediction of potential immunogenic B and T-cell epitopes. Population conservation studies of the selected protein and predicted epitopes were also performed using population conservancy analysis tool of immune epitope database resource. The two dimensional and three dimensional structure of multi-epitope vaccine were predicted and authenticated using PROCheck and Raptor-X servers. Docking results of the multi-epitope vaccine peptide with human leukocyte antigen class I and II alleles predicted efficient binding and the resulted docked models were stable during simulation. In silico immune evaluation using C-ImmSim server showed that the peptide could concurrently elicit cell-mediated and humoral immune responses. Immune simulation studies significantly anticipated high levels of IgM and IgG, T-helper, T-cytotoxic cells, INF-γ.
Keywords
Epitopes, coronavirus, immunoinformatics, bioinformatics, vaccine, docking, COVID-19, SARS-CoV-2
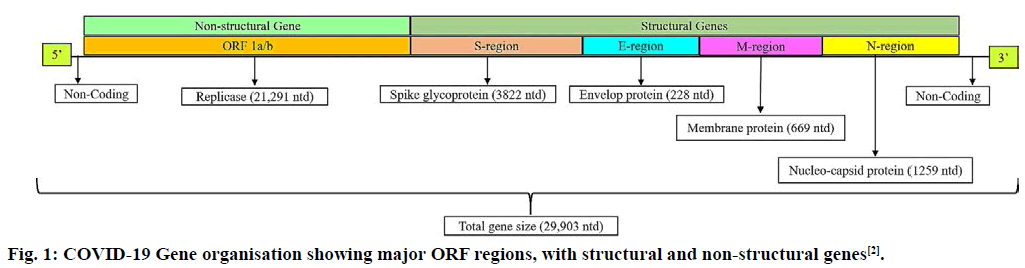
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or Coronavirus disease (COVID-19) is a novel zoonotic virus responsible for outbreak of respiratory illness, which has spread to several countries around the world[1]. The virus was first discovered in Wuhan, China in a 41 y man on 26 December 2019[2]. COVID-19 rapidly triggered a global health emergency, affecting 212 countries by 29 April 2020[3]. Coronaviruses are the members of order Nidovirales, family Coronaviridae and genus β-coronavirus[4]. A total of five major Open Reading Frames (ORF) were found in COVID-19: the ORF 1a/b region coding replicase and other enzymes, the spike or S protein ORF, M or membrane glycoprotein ORF, E or small membrane protein ORF and the N or nucleocapsid ORF (fig. 1)[5]. Its genome consist of single–stranded plus sense RNA which is capped at 5’ end and polyadenylated at 3’ end[6,7]. Contagious human coronavirus (SARS-CoV-2) binds to the receptors of human host cells by transmembrane spike glycoprotein that makes homotrimers protruding from viral surface region[9,10]. Spike glycoprotein has two subunits, S1-responsible for its binding to host cell membrane receptors and S2-responsible for the fusion of viral and host cellular membrane[8,11]. SARS-CoV-2 uses angiotensin converting enzyme-2 (ACE-2) receptor, for entry in human host[11,12]. Serine protease present on cell membrane, is used by SARS-CoV-2 for associating spike protein to enable its fusion with host cells[12-14]. At present no specific drug or vaccine is available for its cure. With an objective of a potent in silico multiepitope vaccine for COVID-19, was designed in this research article. The initial stair of vaccine development process is selecting those antigens that can be used as immunogens. The spike glycoprotein sequence was retrieved from protein data bank (PDB) database. The S1 subunit (PDB I.D-6VSB) of the spike protein being reported to acts as receptor binding domain (RBD), binding to ACE-2 receptor with higher affinity. The T and B cell epitopes were predicted for the S1 subunit protein sequence using Immune Epitope Database (IEDB) server[15].
Figure 1: COVID-19 Gene organisation showing major ORF regions, with structural and non-structural genes[2].
Materials and Methods
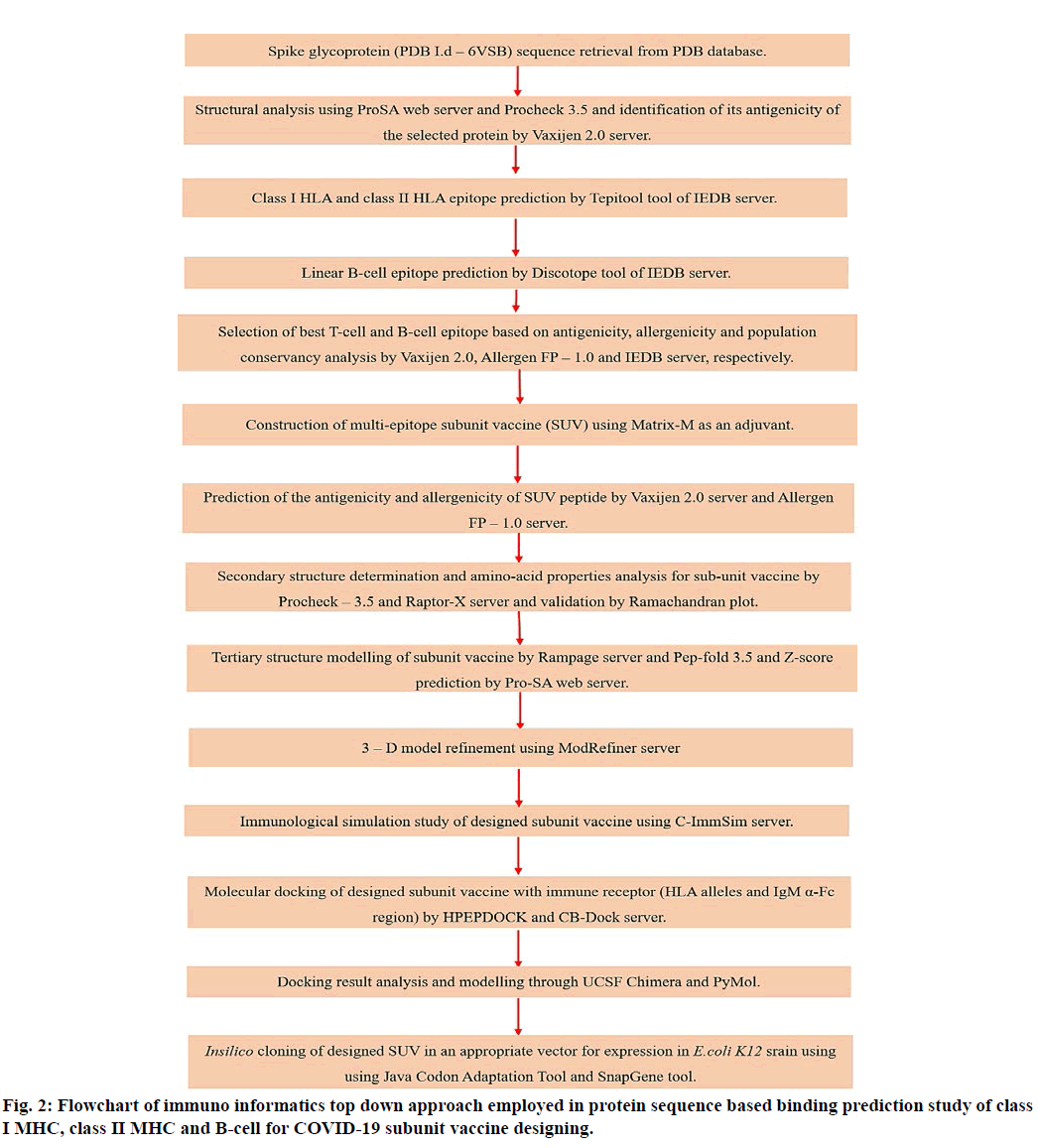
IEDB server was used to select CD8+ T cell (Tc), CD4+ T cell (Th), and B cell epitopes, in accordance with sequence conservation measures covering majorly affected countries population. For the prediction of class I and II HLA (Human Leukocyte Antigen) epitopes Tepitool tool of IEDB server was used. For prediction of B cell epitopes Discotop 1.1 tool of IEDB server was used. All the epitopes selected were validated for their antigenicity and allergenicity by Vaxijen and Allergen FP server, respectively[16]. Further by using these epitopes and an adjuvant the subunit vaccine was designed and evaluated employing.
The subunit vaccine model was constructed and its two dimensional (2D) and three dimensional (3D) structures were predicted and validated using PSIpred, Raptor-X, Procheck 3.5 and Pro-SA web servers respectively[17-21]. The docking studies was performed using CB-dock and HPEPDOCK docking servers to understand the binding capacity between constructed vaccine peptide (ligand) and its protein (receptor)[22-24]. Immune simulation studies were conducted using C-ImmSim server to understand that the multi-epitope vaccine model could elicit IgG and IgM response with CD8+ and CD4+ and T cell mediated immune responses[25]. For the designed vaccine model in silico expression studies on E.coli K12 strain was also performed using SnapGene tool[26].
Immune adjuvants are also used which are unique in their activity and is a key requirement in vaccine formulation. Alhydrogel (Adjuvant I.d-VO-0001241) retrieved from the Vaxjo server was used as an adjuvant for the immune interaction[50]. Alhydrogel is a typical formulation of aluminium hydroxide (Al(OH)3) gels, for immunological investigations[27]. The utilization of aluminium adjuvants is linked with incitement of IL-4 and Th cells, with improved IgG1 and IgE formation[28]. Regarding safety purpose also, Alhydrogel has substantial safety record when used with other recombinant proteins and well tolerated by humans, generating considerable immune responses[29].
Other adjuvant Matrix-M trigger robust immune responses by activating both Th and Tc cell types with low antigen dosages and have a lower risk for allergic responses. Matrix-M is made by saponin, phospholipid and synthetic cholesterol. It can be voluntarily formulated with variety of vaccine and is well-tolerated by humans[30]. The Matrix-M has already been used to cure Ebola pandemic[31,32].
For docking study CB–Dock (http://cao.labshare.cn/ cb–dock/) and HPEPDOCK servers were used. The CB-Dock server forecasts binding sites of provided protein and calculate the centers and sizes using curvature-based cavity detection method and completes docking using Autodock Vina[33]. The CB-Dock prediction server algorithms were sensibly improved and attained >70 % success rate and also gave an interactive 3D visualization of results. The CB-Dock also perform blind docking at predicted sites, instead of whole surface of protein. The HPEPDOCK server executes blind protein-ligand docking using hierarchical algorithm[34,35]. HPEPDOCK also examine the peptide (ligand) flexibility through a group of its conformations created by MODPEP program[36,37].
The reverse transcription and optimization of codon was performed using Java Codon Adaptation Tool (JCat) server to express the multi-epitope vaccine construct in an expression vector[38]. Codon advancement (optimization) was executed to express the finalized vaccine construct in the E. coli K12 strain (host). The codon sequence was optimized by using additional options like, avoiding rho-independent transcription termination, restriction enzymes cleavage sites and prokaryote ribosome binding site. The conceptual methodology employed in designing multi-epitope subunit vaccine for SARS-CoV-2 is given in fig. 2.
Results and Discussion
The complete protein sequences of SARS-CoV-2 (Genbank Accession no. YP_009724390.1) was obtained from PDB (6VSB). The sequence were then subjected to epitope prediction for B–cell and T-cell using IEDB server.
Spike structural glycoprotein of SARS-CoV-2 was analysed for identification of possible human leukocyte antigen (HLA) class-I binding T cell epitopes using Tepitool tool of IDEB server[39,40]. The sequence of spike glycoprotein was screened against 27 most frequent A and B alleles (A*01:01, A*02:01, A*02:03, A*02:06, A*03:01, A*11:01, A*23:01, A*24:02, A*26:01, A*30:01, A*30:02, A*31:01, A*32:01, A*33:01, A*68:01, A*68:02, B*07:02, B*08:01, B*15:01, B*35:01, B*40:01, B*44:02, B*44:03, B*51:01, B*53:01, B*57:01, B*58:01) of class-I HLA. The parameters were customized for removing duplicate peptides, synthesizing 9-mer oligopeptide sequences. The peptides were carefully chosen based on their percentile score and 100 % population conservancy (Table 1). Among 323, 9-mer oligopeptides obtained, a total of 11 potent oligopeptides were selected (Table 2) based on their allergenicity and antigenicity. The allergenicty of selected epitopes were cross-checked by AllerCatPro 1.7 and two HLA class I epitopes found allergic, was discarded. Finally, 9 potent peptides binding to class I HLA was selected for further study.
| Population/Area | MHC Class Combined | ||
|---|---|---|---|
| Population Coverage | Average No. of Hits | PC(90) | |
| Brazil | 100.0 % | 24.78 | 15.38 |
| China | 99.97 % | 21.96 | 13.21 |
| Germany | 100.0 % | 31.32 | 22.32 |
| India | 99.97 % | 20.11 | 10.0 |
| Italy | 99.97 % | 26.52 | 15.22 |
| North America | 100.0 % | 29.17 | 17.69 |
| Spain | 100.0 % | 20.03 | 8.79 |
| Average | 99.99 | 24.84 | 14.66 |
| Standard Deviation | 0.01 | 4.09 | 4.26 |
PC is Population Coverage. MHC is Major Histocompatibility Complex.
Table 1: Population Coverage Calculation Result Obtained By Combining Both MHC Classes
| Epitope Code | Type of Epitope | Peptide Sequence | Start | End | Allergenicity Score |
Antigenicity Score (>0.5) |
|---|---|---|---|---|---|---|
| B–1 | B–cell | VRQIAPGQTGKIAD | 407 | 421 | Non–Allergen | 1.260 |
| B–2 | B–cell | KNHTSPDVDLG | 1156 | 1167 | Non–Allergen | 1.403 |
| MH–1 | MHC II | GYQPYRVVVLSFELL | 504 | 518 | Non–Allergen | 1.074 |
| MH–2 | MHC II | PTNFTISVTTEILPV | 715 | 729 | Non–Allergen | 1.134 |
| MC–1 | MHC I | DVNCTEVPV | 614 | 622 | Non–Allergen | 1.239 |
| MC–2 | MHC I | FLHVTYVPA | 1062 | 1070 | Allergen/ Non–Allergen | 1.334 |
| MC–3 | MHC I | FPNITNLCP | 329 | 337 | Allergen/ Non–Allergen | 1.621 |
| MC–4 | MHC I | VVFLHVTYV | 1060 | 1068 | Non–Allergen | 1.512 |
| MC–5 | MHC I | DIADTTDAV | 568 | 576 | Non–Allergen | 1.090 |
| MC–6 | MHC I | VLSFELLHA | 512 | 520 | Non–Allergen | 1.077 |
| MC–7 | MHC I | SKRVDFCGK | 1037 | 1045 | Non–Allergen | 1.732 |
| MC–8 | MHC I | LEILDITPC | 582 | 590 | Allergen/ Non–Allergen | 1.639 |
| MC–9 | MHC I | LPIGINITR | 229 | 237 | Non–Allergen | 1.821 |
MHC is Major Histocompatibility Complex, which are of two types Major Histocompatibility Complex-I and Major Histocompatibility Complex-II.
Table 2: Predicted B–Cell and MHC Class I and II Epitopes with other Details
The Tepitool tool of IEDB server was used to predict peptides binding to HLA class II[39,40]. Spike glycoprotein sequence obtained from PDB was screened for the panel of 26 most frequent alleles for promiscuous binding (HLA-DPA1*01/DPB1*04:01, HLA-DPA1*01:03/DPB1*02:01, HLA-DPA1*02:01/DPB1*01:01, HLA-DPA1*02:01/DPB1*05:01, HLA-DPA1*03:01/ DPB1*04:02, HLA-DQA1*01:01/DQB1*05:01, HLADQA1* 01:02/DQB1*06:02, HLA-DQA1*03:01/ DQB1*03:02, HLA-DQA1*04:01/DQB1*04:02, HLA-DQA1*05:01/DQB1*02:01, HLA-DQA1*05:01/ DQB1*03:01, HLA-DRB1*01:01, HLA-DRB1*03:01, HLA-DRB1*04:01, HLA-DRB1*04:05, HLADRB1* 07:01, HLA-DRB1*08:02, HLA-DRB1*09:01, HLA-DRB1*11:01, HLA-DRB1*12:01, HLADRB1* 13:02, HLA-DRB1*15:01, HLA-DRB3*01:01, HLA-DRB3*02:02, HLA-DRB4*01:01, HLADRB5* 01:01).
The parameters were customized for removing duplicate peptides, synthesizing 15-mer peptide sequences. Among 211, 15mer peptides obtained, two potent peptides were selected based on percentile score and 100 % population conservancy (Table 2). Linear epitopes of SARS-CoV-2 spike glycoprotein for B-cell was predicted using Discotope 1.1 prediction tool of IEDB[41,42]. The B-cell epitope prediction by Discotope tool integrates solvent-reachable surface area assessment and contact distances to predict B–cell epitope[43]. The predicted epitopes were selected over a threshold value of -7.7 from Discotop 1.1 tool. The two selected epitopes were also analysed for their allergenicity (Allergen FP-1.0 server), antigenicity (Vaxijen 2.0 server) and population conservancy analysis (Table 1). The allergenicty of selected epitopes was cross-checked by AllerCatPro 1.7.
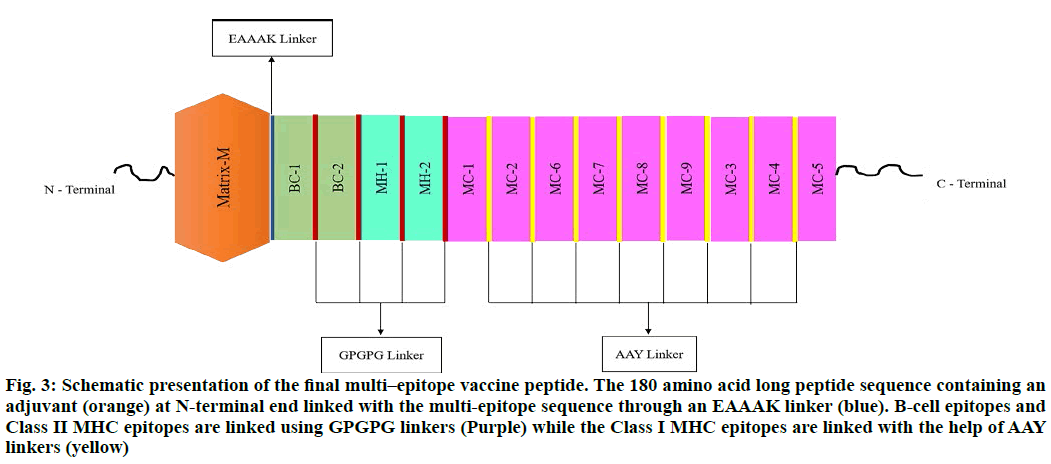
The selected epitopes used in designing the Subunit Vaccine (SUV) were 02 linear B-cell epitopes, 09 major histocompatibility complex (MHC) class I epitopes, and 02 MHC class II epitopes, linked by GPGPG and AAY linkers. The Matrix-M, was opted as an adjuvant and added to the N-terminal of SUV using EAAAK linker. The different orientations of peptides were applied in SUV designing. The final vaccine peptide model of 180 amino acid residues was constructed from 13 merged epitope sequences (fig. 3).
Figure 3: Schematic presentation of the final multi–epitope vaccine peptide. The 180 amino acid long peptide sequence containing an adjuvant (orange) at N-terminal end linked with the multi-epitope sequence through an EAAAK linker (blue). B-cell epitopes and Class II MHC epitopes are linked using GPGPG linkers (Purple) while the Class I MHC epitopes are linked with the help of AAY linkers (yellow)
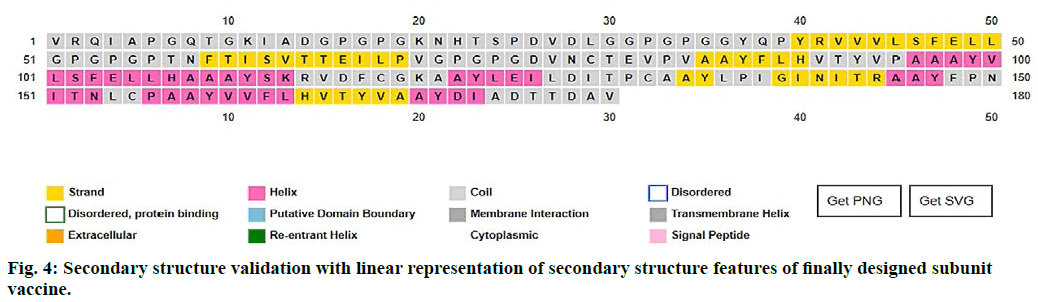
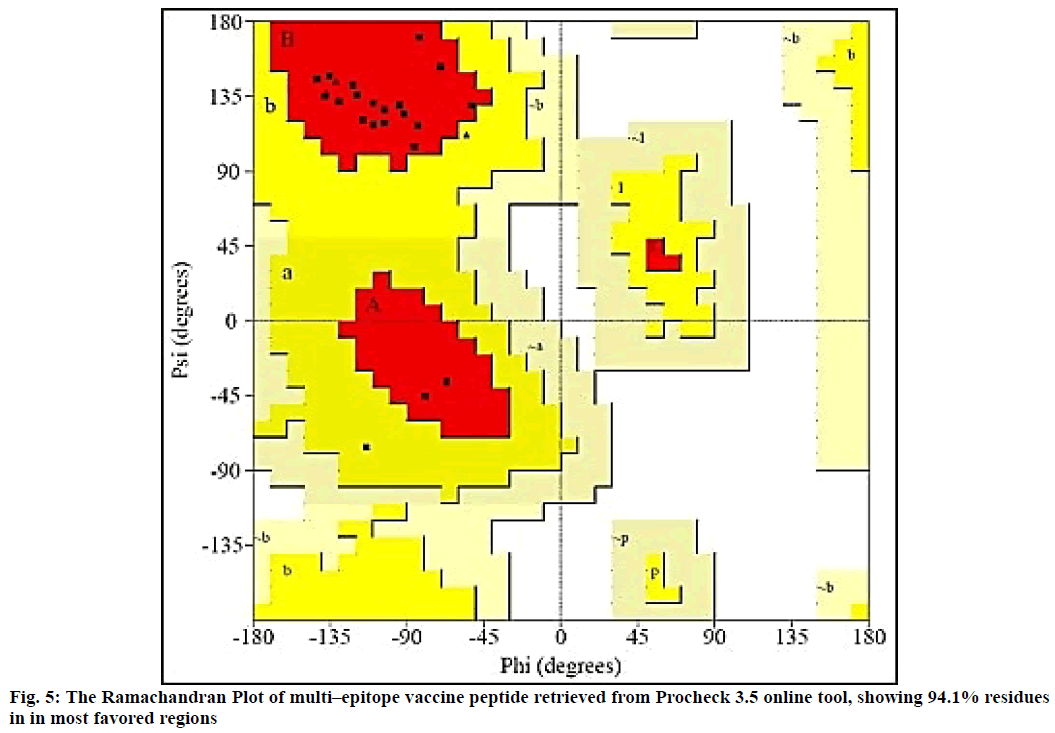
The 2D structure and other parameters of final peptide (SUV) were obtained through PSIPred server, fig. 4. The designed sequence was also analyzed through Raptor-X property prediction tool. The Raptor-X result predicted 93 % positions of amino acids in allowed regions having alpha-helix (22 %), beta-sheet (23 %) and coiled (53 %) structure, respectively. The ramachandran (RMC) plot analysis using Procheck 3.5, resulted 94.1 % amino acid fragments are in favoured regions (A, B, L) and 5.9 % in additionally permitted regions (a, b, l, p) including glycine (4) and proline (4) residues. Based on the analysis of 118 structures of resolution (2.0 Å) and R–factor not more than 20 %, a good calibre (quality) model was expected, having more than 90 % residues in favoured regions (fig. 5).
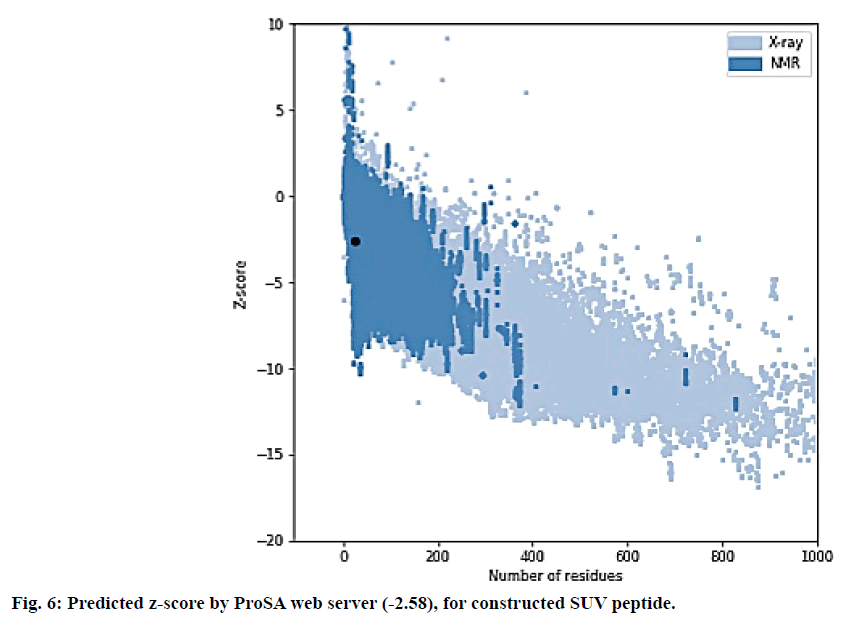
The SUV peptide sequence generated was subjected for 3D structure modelling by PEPfold-3 and Swiss-Model. The Ramachandran (RMC) plot to predict the validity of 3D structure of final peptide was analysed form Procheck 3.5[44-46]. The 3D structure of final peptide was also validated through RAMPAGE and the result depicted 95.8 % residues in favoured region. The peptide of final vaccine subunit indicated decent alignment as per Z-score (fig. 6) value (reaching between 1.18 and 5.46) through ProSA-web server. The ProSA-web gave a Z-score of -2.58. The finally validated and selected model after refinement through ModRefiner server had overall quality factor of 89 %.
C-ImmSim (immune simulation) server yield reliable results with actual immune responses. The various antigen-antibody interacttions are shown in fig. S1(a) (supplementary file). The primary response was depicted by significant rise in levels of IgG and IgM. The secondary and tertiary responses represented marked increments in B-cell, including levels of IgG1 and IgG2, IgM, and IgG+IgM antibodies with a corresponding decrease in antigen concentration, fig. S1(b), S1(c) (supplementary file). Similarly, high response was observed in the Th and Tc cell population with corresponding memory development (fig. S1(c) and S1(d) supplementary file). The SUV peptide exposure elicited levels of IgG1, IFN-γ and Th cell concentration.
The binding affinity of SUV peptide was detected with IgM (α-Fc receptor), class I and II HLA. For docking purpose, the α-Fc receptor sequence of IgM was retrieved from genbank (Accession No-NP_001116451.1) and secondary structure was constructed using Swiss Model online server (https://swissmodel.expasy.org/ interactive). The constructed model was retrieved in PDB format and refined using ModRefiner tool (RMSD score - 1.432 and Tm score-0.9486). The protein sequences of class I HLA (Accession No-CCH75804.1) and class II HLA (Accession No-QCF28413.1) was also retrieved from Genbank and secondary structure was constructed for docking study. The final structure of IgM (α-Fc receptor), class I HLA and class II HLA was refined by ModRefiner tool and analysed by using UCSF chimera.
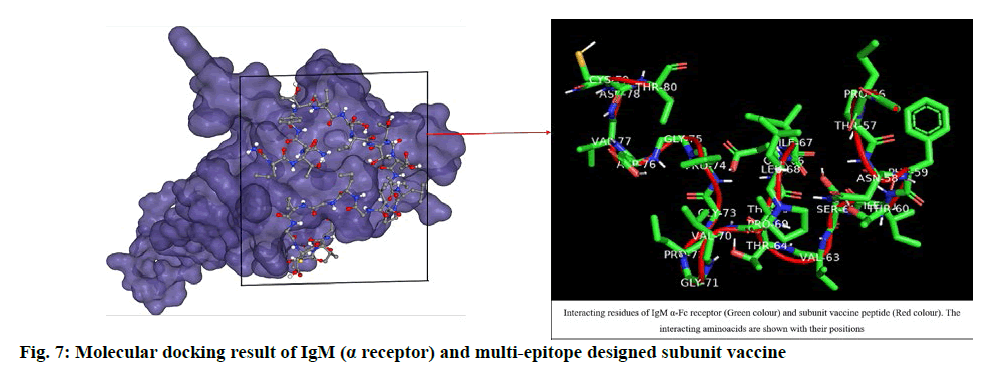
The designed SUV peptide was used as ligand and IgM (α-Fc receptor) was used as receptor to perform blind docking simulation. The result of HPEPDock server depicted its top model with -182.84 docking score and the CB-Dock server gave -5.5 vina score and 401 Å cavity size volume of receptor molecule. The respective result (fig. 7) obtained was analysed by using Pymol. The docking score (binding affinity) revealed that the ligand and receptor interaction is stable enough to generate immune responses.
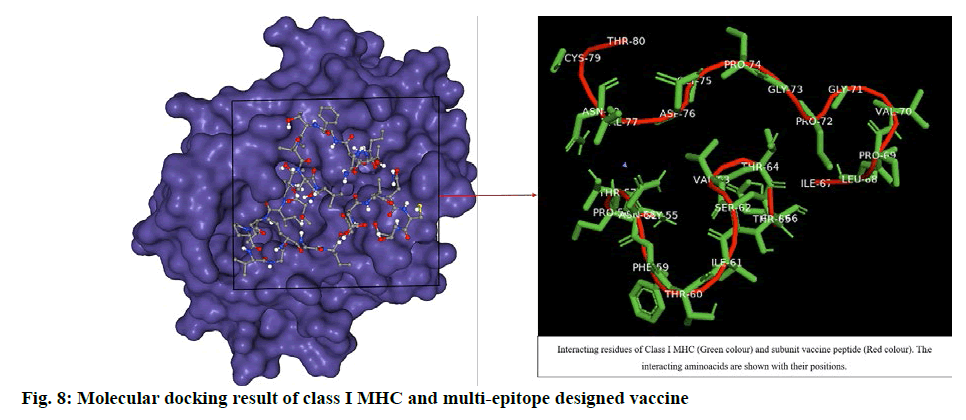
The ligand was blindly docked with class I HLA protein and the result depicted -235.40 docking score from HPEPDOCK server top model and -5.6 Vina score with 320 Å cavity size volume of receptor molecule for CB-Dock top model (Table S2, supplementary file). The respective result (fig. 8) obtained was analysed by using UCSF Chimera[47]. The docking score revealed that the ligand and receptor interaction was stable enough to generate immune responses, activating Tc cells and release of IFNγ.
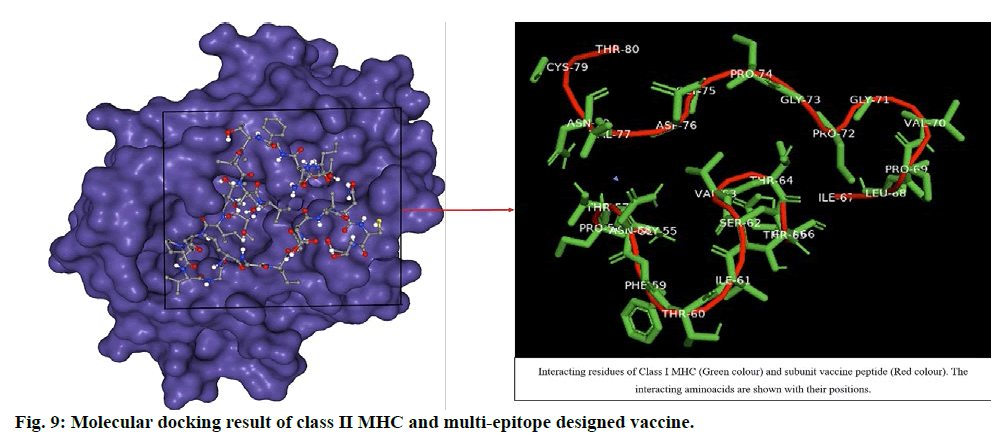
The ligand was blindly docked with class II MHC and the result depicted -286.78 docking score from HPEPDOCK server top model and -7.0 Vina score with 249 Å cavity size volume of receptor molecule for CB-Dock top model (Table S2, supplementary file). The respective result (fig. 9) obtained was analysed by using Pymol. The docking score revealed that the ligand and receptor interaction is stable enough to generate immune responses and develop a good response through class II MHC activating Th cells and release of interleukins.
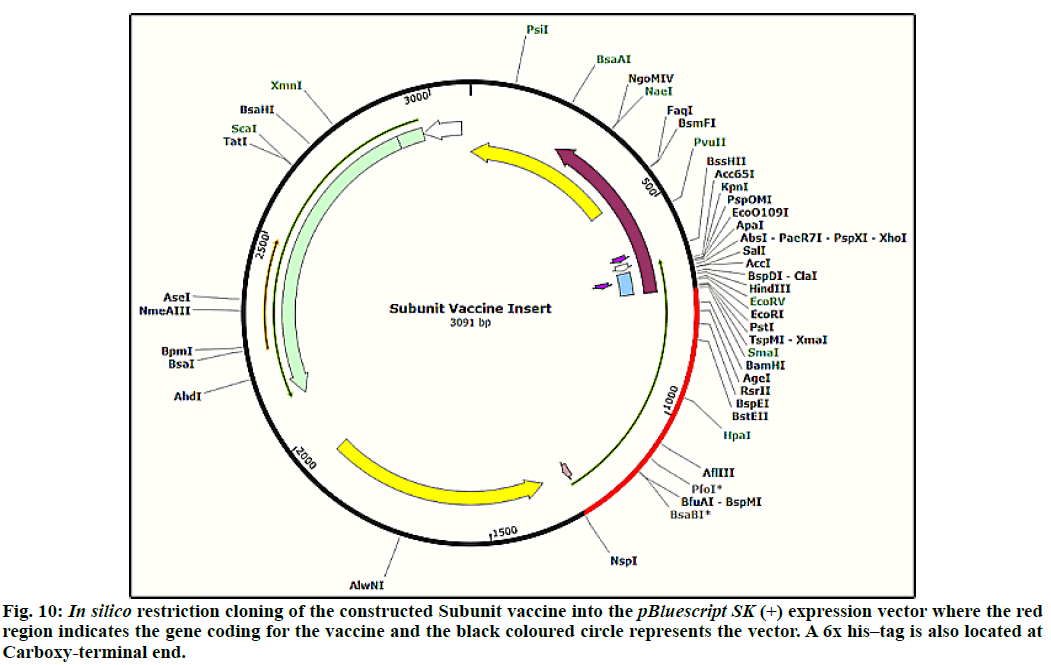
The JCat tool resulted CAI (codon adaptation index) as 1.0 and percentage GC content as 56.66, which were used to evaluate expression level of protein[48]. The CAI value of 0.8 to 1.0 indicates a good score and limits codon usage partialities. In order to clone final optimized subunit vaccine peptide construct in E. coli, pBluescript SK(+) vector of 2958 bp was used. Restriction sites of Bam HI and Nsp I were inserted at N and C-terminals of gene sequence, respectively. Before Nsp I restriction site a 6x histidine tag was also added to aid the purification methods. Finally, the optimized sequence was ligated into the pBluescript SK(+) vector using SnapGene tool to confirm vaccine expression (fig. 10). The size of final clone constructed was 3091 base pairs.
The COVID-19, as named by WHO (World Health Organisation), is a contagious virus which created havoc death situations around the world. More than 3.5 million affected cases and above two million deaths have occurred around the world till 30th December 2020[51,52]. For the present research work, spike glycoprotein sequence (PDB I.d-6VSB) was retrieved from PDB database. The subsequent protein sequence was then used to identify potential epitopes (B-cell and T-cell). The B-cell activation is responsible for the outcome of humoral immune response and also the development of memory. The IgM with IgG generates humoral immune response and also acts as antigen presenting cells (APCs). The B-cell epitopes which were determined by IEDB prediction server have the potential to bring out the activation of IgG and IgM which are necessary to develop humoral response and memory cells in the body. The selected B–cell epitopes based on their propensity score was then subjected to check their allergenicity and antigenicity by Vaxijen 2.0 and Allergen FP 1.0 online server (Table 1). For prediction of T-cell epitopes for class I and II HLA Tepitool tool of IEDB server was used and epitopes were selected based on their IC50 value. The selected HLA epitopes were then identified for their allergenicity and antigenicity over a threshold value of 0.7 (Table 2). As the HLA allele differs in distribution among diverse geographical regions and ethnic clusters around the world. Population coverage analysis was performed and major population coverage (99 to 100 %) was perceived for particular 13 epitopes in majorly affected countries (Table 1).
Among the two selected adjuvants, Matrix-M is newly introduced adjuvant and reported to be used in various viral vaccine developments. It has the potential to give both B and T-cell facilitated better immune response. Now a days Matrix-M is being frequently used to design subunit multi-epitope vaccines[49]. As per the research, Matrix-M was proved to be more antigenic then alhydrogel[49]. The B and T-cell epitopes binding to their specified HLA alleles are shown in Table S1, supplementary file. The subunit vaccine model was constructed using class I HLA (9), class II HLA (2) and B-cell (2) epitopes by using AAY and GPGPG linkers.
The Ramachandran plot (fig. 5) depicted that 96.5 % residues were in favorable region and the Z–score predicted by ProSA server was -2.58. The Z-score validated that the vaccine peptide could be constructed and resulting better immunological response. The immune simulation results predicted that the designed vaccine model was potent enough to activate B-cell associated IgG and IgM response, among which IgG was able to develop memory of the encounter and can be preserved in the body for secondary response. The subunit vaccine was also potent to develop class I and II MHC response. The vaccine peptide also activated IFN-γ, IL-17, IL-26 and macrophages, involved in both adaptive and innate immunity.
The docking studies predicted strong binding affinities with the vaccine peptide and their receptors giving a dock score of -5.6, -7.0 and -5.5 (Table S2, supplementary file) respectively. The docking results were further analysed by Pymol and interacting residues are depicted in fig. 7-9. The cloning studies of SUV peptide reveal its better integration (fig. 10) for the expression in E.coli K12 strain.
Acknowledgements:
The author is thankful to Dr. Gajendra B. Singh, Associate Professor, University Institute of Biotechnology, Chandigarh University, Punjab.
References
- Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-an update on the status. Military Med Res 2020;7(1):1-10.
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nat 2020;579(7798):265-269.
- World Health Organization. Coronavirus disease 2019 (COVID-19): situation report 2020;72.
- Pal M, Berhanu G, Desalegn C, Kandi V. Severe Acute Respiratory Syndrome Coronavirus–2 (SARS–CoV–2): An Update. Cureus 2020;12(3): e7423.
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181(2):282-92.
- Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J 2019;16(1):69-80.
- Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID–19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res 2020;24:91-8.
- Kawase M, Kataoka M, Shirato K, Matsuyama S. Biochemical analysis of coronavirus spike glycoprotein conformational intermediates during membrane fusion. J Virol 2019;93(19):785-819.
- Hoffmann M, Kleine Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181(2):271-80.
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L. Characterization of spike glycoprotein of SARS–CoV–2 on virus entry and its immune cross–reactivity with SARS-CoV. Nat Comm 2020;11(1):1-12.
- Zhuang M, W Cheng, Y Zhang, J Jiang, X M, Wang L, Deng J, et al. Increasing host cellular receptor—angiotensin?converting enzyme 2 expression by coronavirus may facilitate 2019?nCoV (or SARS?CoV?2) infection. J med virol 2020;92(11):2693-701.
- Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nat 2013;500(7461):227-31.
- Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sinica 2020;41(9):1141-9.
- Rahman N, Basharat Z, Yousuf M, Castaldo G, Rastrelli L, Khan H. Virtual screening of natural products against Type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2). Molecules 2020;25(10):2271-83.
- Astuti I, Ysrafil. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS–CoV–2): An overview of viral structure and host response. Diabet Metabol Syndrome 2020;14(4):407-12.
- Joshi A, Joshi BC, Mannan MAU, Kaushik V. Epitope based vaccine prediction for SARS-COV-2 by deploying immuno-informatics approach. Inform Med Unlock 2020;19:100338-52.
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallo 1993;26(2):283-91.
- Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK–NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 1996;8(4):477-86.
- Wiederstein M, Sippl MJ. ProSA–web: interactive web service for the recognition of errors in three–dimensional structures of proteins Nucleic acids res 2007;35(suppl-2):W407-W410.
- Sippl MJ. Recognition of errors in three?dimensional structures of proteins. Proteins 1993;17(4):355-62.
- Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins 1992;12(4):345-64.
- Abd SA, Al-Nour M, Elhag M, Idris AA, Haroun E, Adam ME, et al. A Multiple Peptides Vaccine against COVID-19 Designed from the Nucleocapsid phosphoprotein (N) and Spike Glycoprotein (S) via the Immunoinformatics Approach. Inform med unlock 2020;21:100476-100489.
- Çak?r B, Okuyan B, ?ener G, Tunali-Akbay T. Investigation of beta-lactoglobulin derived bioactive peptides against SARS-CoV-2 (COVID-19): In silico analysis. Eur J Pharmacol 2020;891:173781-96.
- Liscano Y, Oñate-Garzón J, Ocampo-Ibáñez ID. In Silico Discovery of Antimicrobial Peptides as an Alternative to Control SARS-CoV-2. Molecules 2020;25(23):5535-48.
- Bohra N, Sasidharan S, Raj S, Balaji SN, Saudagar P. Utilising capsid proteins of poliovirus to design a multi-epitope based subunit vaccine by immunoinformatics approach. Mol Simul 2020;46(5):419-28.
- Kalita P, Padhi AK, Zhang KY, Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microbe Pathogen 145;104236-52.
- Agarwal S, Hickey JM, McAdams D, White JA, Sitrin, R, Khandke L, et al. Effect of aluminum adjuvant and preservatives on structural integrity and physicochemical stability profiles of three recombinant subunit rotavirus vaccine antigens. J Pharma Sci 2020;109(1):476-87.
- Eisenbarth SC, Colegio OR, O’Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nat 2008;453(7198):1122-6.
- Munks MW, McKee AS, MacLeod MK, Powell RL, Degen JL, Reisdorph NA, et al. Aluminum adjuvants elicit fibrin–dependent extracellular traps in vivo Blood. J American Soc Hematol 2010;116(24): 5191–5199.
- Cox F, Baart M, Huizingh J, Tolboom J, Dekking L, Goudsmit J, et al. Protection against H5N1 influenza virus induced by matrix–M adjuvanted seasonal virosomal vaccine in mice requires both antibodies and T cells. PloS one 2015;10(12):1-15.
- Bengtsson KL, Song H, Stertman L, Liu Y, Flyer DC, Massare MJ, et al. Matrix–M adjuvant enhances antibody, cellular and protective immune responses of a Zaire Ebola/Makona virus glycoprotein (GP) nanoparticle vaccine in mice. Vaccine 2016;34(16):1927-35.
- Suschak JJ, Schmaljohn CS. Vaccines against Ebola virus and Marburg virus: recent advances and promising candidates. Human Vacc Immunothera 2019;15(10):2359-77.
- Zhou P, Jin B, Li H, Huang SY. HPEPDOCK: a web server for blind peptide–protein docking based on a hierarchical algorithm. Nucleic acids Res 2018;46(W1):W443-50.
- Huang SY, Zou, X. An iterative knowledge?based scoring function for protein-protein recognition Proteins 2008;72(2):557-79.
- Yan Y, Zhang D, Huang SY. Efficient conformational ensemble generation of protein–bound peptides J Cheminform 2017;9(1):59-70.
- Huang SY, Zou X. Ensemble docking of multiple protein structures: considering protein structural variations in molecular docking. Proteins 2007;66(2):399-421.
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic acids Res 2018;46(W1):W296-303.
- Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic acids Res 2005;33(suppl_2):W526-31.
- Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform 2006;7(1):153-68.
- Cao Y, Li L. Improved protein–ligand binding affinity prediction by using a curvature-dependent surface-area model. Bioinform 2014;30(12):1674-80.
- Kringelum JV, Lundegaard C, Lund O, Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol 2012;8(12):p.e1002829.
- Paul S, Sidney J, Sette A, Peters B. TepiTool: a pipeline for computational prediction of T cell epitope candidates. Curr Prot Immunol 2016;114(1):181911-24.
- Pourseif MM, Parvizpour S, Jafari B, Dehghani J, Naghili B, Omidi Y. A domain-based vaccine construct against SARS-CoV-2, the causative agent of COVID-19 pandemic: development of self-amplifying mRNA and peptide vaccines. BioImpacts 2021;11(1):65-82.
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallo 1993;26(2):283-91.
- Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK–NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 1996;8(4):477-86.
- Wang S, Peng J, Ma J, Xu J. Protein secondary structure prediction using deep convolutional neural fields. Sci Rep 2016;6(1):1-11.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Compute Chem 2004;25(13):1605-12.
- Shankar U, Jain N, Mishra SK, Sk MF, Kar P, Kumar A. Mining of Ebola virus genome for the construction of multi-epitope vaccine to combat its infection. J Biomole Stru Dynam 2021;1-17.
- Ghosh P, Bhakta S, Bhattacharya M, Sharma AR, Sharma G, Lee SS, et al. A Novel Multi-Epitopic Peptide Vaccine Candidate Against Helicobacter pylori: In-Silico Identification, Design, Cloning and Validation Through Molecular Dynamics. Internat J Pep Res Therapeut 2021;1-18.
- Sunita, Sajid A, Singh Y, Shukla P. Computational tools for modern vaccine development. Human Vac Immunotherap 2020;16(3):723-35.
- Waters LJ, Pozniak AL. COVID-19 death in people with HIV: interpret cautiously. The Lancet HIV 2021;8(1):e2-e3.
- Malekhosseini SA, Nikoupour H, Gholami S, Shamsaeefar A, Arasteh P, Kazemi K, et al. A report of 85 cases of COVID-19 and abdominal transplantation from a single center: what are the associated factors with death among organ transplantation patients. Transplantation 2021;105(1):90-9.