Nanobodies: COVID-19 and Future Perspectives
- 1Institute of Medicine, Faculty of Medicine, Universidad Austral de Chile, Valdivia, Chile
- 2School of Medicine, University of Costa, San José, Rica
- 3Institute of Biochemistry and Microbiology, Universidad Austral de Chile, Valdivia, Chile
- 4Berking Biotechnology, Valdivia, Chile
- 5Institute of Philosophy and Complexity Sciences, Santiago, Chile
The COVID-19 pandemic has driven biotechnological developments to provide new and more effective tools for prophylaxis, diagnosis, and therapy. Historically, monoclonal antibodies have been valuable tools; however, the pandemic has shown some weaknesses, such as production limitations at a global scale. An alternative to conventional monoclonal antibodies are nanobodies, recombinant fragments of the variable region of single-domain antibodies derived mainly from the Camelidae family. Nanobodies have multiple characteristic benefits: they are small (15 KDa) and have remarkable refolding capability and unlimited possibilities for modifications due to their recombinant nature. Here, we review the application of nanobodies in diagnosis and treatment of SARS-CoV-2 infection.
Introduction
Heavy-chain-only antibodies (HCAbs) are found in camelids (e.g., dromedaries, camels, llamas, and alpacas) (Arbabi-Ghahroudi, 2017) and some cartilaginous fish species such as nurse shark (Ginglymostoma cirratum), wobbegong shark (Orectolobus maculatus), spiny dogfish (Squalus acanthias), and smooth dogfish (Mustelus canis) (Camacho-Villegas et al., 2013; Cheong et al., 2020; Dooley and Flajnik, 2006; Dooley et al., 2006; Feige et al., 2014; Ohtani et al., 2013; Roux et al., 1998; Stanfield et al., 2004; Nuttall et al., 2001). Within their immune system, type IgG2 and IgG3 lack the CH1 domain due to alternative splicing. Consequently, the light chains do not pair to the final antibody leading to the generation of HCAbs (Hamers-Casterman et al., 1993). IgG2 and IgG3 rely on heavy variable domains or VHH, a small domain of approximately 15 kDa to bind to specific antigens (Muyldermans et al., 1994; Desmyter et al., 1996; Arbabi Ghahroudi et al., 1997; Vu et al., 1997; Hassanzadeh GH et al., 1998; Lauwereys et al., 1998). The key difference between conventional monoclonal antibodies and HCAbs is that the latter are encoded from single genes (Figure 1A). Thus, the VHH can be amplified from cDNA, cloned into libraries of diverse natures, and later isolated using conventional molecular biology procedures. Once isolated, the recombinant VHH fragments of HCAbs are known as nanobodies (Muyldermans et al., 1994). Nanobodies conserve selective binding to specific antigens in the ∼15 KDa minimal structure, 10 times smaller than conventional antibodies (Arbabi Ghahroudi et al., 1997). Nanobodies are soluble and highly stable to a wide range of pH and temperatures (Bond et al., 2003), along with an outstanding penetrability (Muyldermans, 2013) (Figure 1B). Some nanobodies are able to penetrate the blood-brain barrier which places them as promising tools for future diagnosis and therapy for neurodegenerative diseases. Among all the outstanding advantages of nanobodies, one of the most attractive is the performance achieved through recombinant expression systems. Nanobodies can be produced efficiently in eukaryotic and prokaryotic systems (Arbabi-Ghahroudi et al., 2005; Liu and Huang, 2018; de Marco, 2020) and, because nanobody sequences are obtained during the selection process, the final molecule can be engineered to cope with several applications using conventional molecular biology tools. Moreover, open-source computational pipelines are available for nanobody humanization such as “Llamanade” (Sang et al., 2022).
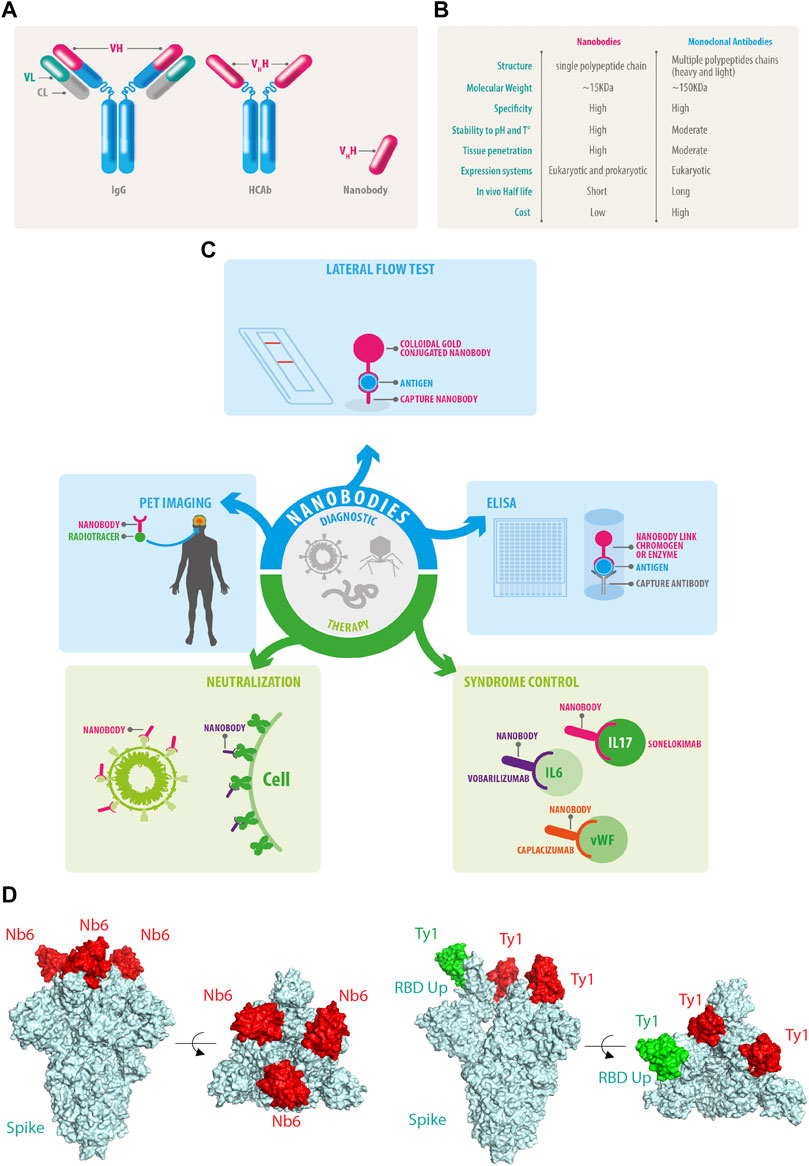
FIGURE 1. Nanobody features and functions. (A) Conventional IgG antibodies contain heavy and light chains, HCAb does not associate with light chains allowing the isolation of the genetic fragments of the Vhh also known as nanobodies. (B) Comparative features of nanobodies versus monoclonal antibodies. (C) Current and future perspectives of nanobodies. (D) Nanobodies bind to the RBD of SARS-CoV-2 and neutralize the infection by different mechanisms: NB6 binds to the RBD of each Spike protomer in a down conformation (PDB ID: 7KKK), while Ty1 allows the upper conformation of one RBD of Spike in green (PDB ID: 6ZXN), in both examples, the nanobodies prevent the engagement of the human ACE2 receptor.
Nanobodies are remarkable tools for diagnosing and treating various diseases, and the COVID-19 pandemic has driven the generation of several nanobodies against SARS-CoV-2. Here, we summarize some of the current applications and production strategies of nanobodies to help fight viral pathogens.
Nanobody Expression Systems
Nanobodies are compact structures and can be produced in high yields in several expression systems, such as bacteria, yeast, mammalian cells, and plants (Frenken et al., 2000; Arbabi-Ghahroudi et al., 2005; Ismaili et al., 2006).
Production in prokaryotes has a low cost and involves easy handling (Rosano et al., 2019). The traditional way to express recombinant proteins in bacteria is in the cytoplasm; however, the reducing cytoplasmic conditions can negatively affect the formation of disulfide bonds in some nanobodies, which are necessary for the correct folding of its tertiary structure (Govaert et al., 2012; Muyldermans, 2013; Hagihara and Saerens, 2014; Billen et al., 2017). Despite this, there are examples of nanobodies produced under cytoplasmatic conditions, which require the attachment of other proteins (Anderson et al., 2018), or expression in special bacterial strains with an oxidizing cytoplasmatic environment such as SHuffle T7 cells (Li et al., 2019) or co-expression with enzymes such as Erv1p sulfhydryl oxidase (Veggiani and de Marco, 2011; Shriver-Lake et al., 2017). High yields are also obtained when expressing proteins in classical inclusion bodies and further by denaturation in urea-mediated protein extraction (Maggi and Scotti, 2017). Also, there is an interesting alternative of expressing the nanobody coupled to a secretion pathway, such as the hemolysin secretion system (Günaydın et al., 2014; Ruano-Gallego et al., 2019).
Nowadays, the most convenient method for low-scale production of recombinant nanobodies is in Escherichia coli periplasm (Salema et al., 2013). Its oxidizing environment ensures correct folding and disulfide bond formation, and the periplasmic extracts enriched with the recombinant nanobodies facilitate subsequent purification (Conrath et al., 2001; Billen et al., 2017). The N-terminal pelB leader sequence drives the protein to the post-translational Sec pathway, the nanobodies are completely synthesized in the ribosome and then released to the Sec-translocase, which enables its carriage through the inner bacterial membrane and accumulates the nanobodies in the periplasm (Keen and Tamaki, 1986; Yoon et al., 2010; Billen et al., 2017). The recombinant proteins that accumulate in the periplasmic space are usually recovered after an osmotic shock which softly breaks the outer bacterial membrane, allowing the release of the proteins and preventing further contamination with E. coli cytoplasmatic proteins. Affinity purification can be applied directly after using various affinity matrices, such as hexahistidine (His6)-tag (Salema and Fernández, 2013), maltose-binding protein (MBP), or the Avi-tag, which allows in vivo biotinylation of the tagged protein (Hernot et al., 2012; Zhu et al., 2014; Noor et al., 2018; Sun et al., 2018; Du et al., 2019).
Nanobodies can also recognize linear peptides, an example of this is a nanobody called ALFA Selector, which recognizes the short ALFA-tag (SRLEEELRRRLTE) sequence, a very efficient tag for protein purification, including nanobodies themselves (Götzke et al., 2019; Kilisch et al., 2021).
Human or humanized monoclonal antibodies have been approved during the pandemic as emergency treatment measures. However, the production limitation led to the access of being almost exclusive to high-income countries in the northern hemisphere. The production of conventional antibodies occurs in mammalian cells, which is costly and time-consuming. In most cases, single clones of mammalian HEK293 or CHO cells must be isolated and expanded into cell banks, a process that takes several months. In contrast, therapeutic nanobodies and nanobodies fused to the Fc of conventional antibodies (Nb-Fc) can be produced in yeast, such as Pichia pastoris, in an endotoxin-free manner and at a very low cost. Interesting technologies based on CRISPR/Cas9 improve the glycosylation homogeneity of P. pastoris, which could allow efficient expression for therapeutic technologies using nanobodies (Krainer et al., 2013; Weninger et al., 2016; Schepens et al., 2021).
Nanobodies as Diagnostic Tools
Immunoaffinity techniques are the primary tools for rapid diagnosis, not only in infectious diseases but also in an extensive range of pathologies. Nanobodies are incipient in this field, and some technical difficulties regarding their small size and low retention on nitrocellulose strips remain to be solved. Serological tests have been implemented for parasitic infections such as Taenia solium cysticercosis (Deckers et al., 2009; Huang et al., 2010), Trypanosoma spp (Saerens et al., 2008a), and bacterial diseases, such as S. aureus (Stijlemans et al., 2004; Stijlemans et al., 2017). Indeed, this proof of concept opens up a large field of action for nanobodies in the upcoming years.
Nanobodies have been generated to capture several viral proteins of HIV (Gray et al., 2017); norovirus (Koromyslova and Hansman, 2017), dengue (Fatima et al., 2014) among others. When used for diagnosis, nanobodies can be classified in two groups: the primary nanobodies responsible for the recognition of a pathogen or molecule of interest and the secondary antibodies that bind to primary antibodies and unveil its presence by colorimetric or enzymatic reactions. Nanobodies as recombinant proteins can be modified to accomplish the function of primary and secondary antibodies simultaneously, for instance, nanobody fusion to enzymes such as horseradish peroxidase (HRP) has been used for the detection of anti-Newcastle disease virus (NDV) antibodies in chicken sera (Sheng et al., 2019).
Significant efforts have been made to develop diagnostic techniques for COVID-19, focusing mainly on speed and accuracy. The first available tools were based on conventional monoclonal IgM and IgG lateral flow immunoassays (Yetisen et al., 2013; Goossens et al., 2017); however, the expression of IgM and IgG against SARS-CoV-2 antigens is only detectable in late disease stages. Therefore, the gold standard diagnostic test for SARS-CoV-2 diagnosis is the reverse-transcription quantitative polymerase chain reaction (RT-qPCR) from nasopharyngeal and oropharyngeal swab samples (Wang et al., 2020a; To et al., 2020). After vaccines were implemented, IgM and IgG lateral flow immunoassays became useful to determine the immune response against vaccines. Recently developed secondary nanobodies showed superior properties for cellular biology studies regarding penetrance, staining accuracy and, furthermore, secondary nanobodies can be premixed with primary antibodies to bypass the primary antibody animal-species limitations (Sograte-Idrissi et al., 2020). Nanobodies can be fused to the Fc of conventional immunoglobulins and produced recombinantly, which complements nanobody binding capabilities with several technologies already available for monoclonal antibodies (Bao et al., 2021; Girt et al., 2021; Valenzuela Nieto et al., 2021) (Figure 1C).
Nowadays nanobody-based lateral flow tests can rapidly detect recombinant human interferon α2b (Qin et al., 2021). They have also been used to detect active Trypanosoma congolense infections (Pinto Torres et al., 2018). In addition to rapid tests, nanobodies are efficient and sensitive tools for enzyme-linked immunosorbent assay (ELISA), a method allowing the capture and quantitative measure of antigens in the small absorbent surface. In short, nanobodies can be immobilized on the surface of ELISA plates to capture the molecule of interest, and further a second non-competitive nanobody associated to enzymatic activity can be applied to detect the already trapped molecule of interest. Nanobodies covalently coupled to HRP provide sensitive detection of SARS-CoV-2-specific full-length trimeric spike or RBD (Valenzuela Nieto et al., 2021).
In recent years, miniaturization of chips and sensing layers for biosensor equipment linked with microfluidic devices have been proposed as the best option to obtain the most sensitive detection level (Conroy et al., 2009). However, biosensor sensitivity depends on the physical properties of the molecule that binds its target (Saerens et al., 2008b). One of the advantages of nanobodies is their small size, allowing higher molecule density on a surface, and the possibility of easy directional immobilization, which translates into a higher ligand binding capacity leading to improved sensitivity for detecting low analyte concentrations (Huang et al., 2005). Another recently developed system is the use of nanobody-based organic electrochemical transistors (OECTs) which applies a conjugated polymer and a nanobody to detect SARS-CoV-2 spike protein (Guo et al., 2021).
In the field of non-infectious diseases, nanobodies outperform some conventional antibodies, for instance, in the detection of human prostate-specific antigen, an early marker of prostate cancer (Huang et al., 2005; Saerens et al., 2005). Nanobodies have also been conjugated with radioisotopes as modern diagnostic tools for personalized imaging medicine such as positron emission tomography (PET) to detect EGFR, a tyrosine kinase receptor that is highly expressed in most epithelial cancer cells, believed to contribute to tumor malignancy (Penault-Llorca et al., 2006; Gainkam et al., 2008; Tijink et al., 2008). PET imaging is a promising and emergent field for nanobody applications in in vivo imaging (Figure 1C). It requires the accumulation of radiolabeled nanobodies at the target tissue or tumor and rapid excretion of the remanent circulating nanobodies to distinguish between the real signal and background (Massoud and Gambhir, 2003). Most radioisotopes used for in vivo imaging have a short half-life; for instance, Fluorine 18F decays by positron (β+) emission with a half-life of 109.7 min. Conventional antibodies persist in circulation, leading to a high background signal for some applications. In contrast, nanobodies are generally cleared rapidly suggesting a potential extended use for PET diagnostics (Tijink et al., 2008; Harmand et al., 2021). Nanobodies are expected to become important for cancer diagnosis: Molecules including monoclonal antibodies (Frigerio et al., 2021) used for PET diagnostics are useful for directed radiotherapies through the simple exchange of radioisotopes with, for instance, Lutetium 77Lu. These radiotracers used for diagnostics and therapeutical applications are known as Theranostics (Debnath et al., 2022; Woźniak et al., 2022).
Nanobodies show plenty of advantages and overcome some of the problems observed with conventional antibodies, creating many opportunities for future diagnostic applications.
Immunotherapies Based on Nanobodies
Previous to the COVID-19 pandemic, nanobody-based therapeutic approaches were developed against viruses of global concern such as human immunodeficiency virus-1 (HIV-1) (Chen et al., 2008; Gong et al., 2012; McCoy et al., 2012; Matz et al., 2013), influenza viruses (Ashour et al., 2015; Schmidt et al., 2016), hepatitis C virus (HCV) (Tarr et al., 2013), respiratory syncytial virus (RSV) (Hultberg et al., 2011; Schepens et al., 2011), and enteric viruses (Wu et al., 2017). Antiviral nanobodies were also tested in clinical trials against rotavirus and the human respiratory syncytial virus (RSV). Remarkably, ALX-0171, a trivalent nanobody that neutralizes RSV, substantially decreased the viral load in children. Additionally, it is the first nanobody-based treatment delivered by nebulization through the airway (Palomo and Mas, 2016; Stohr and Palomo, 2016; Larios Mora and Gallup, 2018). Nanobodies have significant advantages when used as a therapy (Chakravarty et al., 2014; Jovčevska and Muyldermans, 2020). Several nanobodies were developed to modulate the immune and inflammatory responses. For example, vobarilizumab (ALX-0061) (Dörner et al., 2017) is a bispecific anti-IL6-R nanobody which has been engineered to extend its half-life targeting human serum albumin; secukinumab (ALX-0761) is a trivalent nanobody against IL-17A/F (Langley et al., 2014; De Munter et al., 2018; Svecova et al., 2019; Xie et al., 2019); caplacizumab (ALX-0081 or ALX-0681) is a bivalent humanized anti–von Willebrand Factor (vWF) nanobody (Abdelghany and Baggett, 2016; Peyvandi et al., 2016; Peyvandi et al., 2017; Scully et al., 2019), which has received approval from the European Medicines Agency (EMA) and the Food and Drug Administration (FDA) in the United States, for treating patients with thrombotic thrombocytopenic purpura (Jovčevska and Muyldermans, 2020), becoming the first nanobody approved for clinical therapy of a chronic disease (Figure 1C).
The primary global response to the COVID-19 pandemic was the creation of efficient vaccines (Li et al., 2020; Li et al., 2021a; Awadasseid et al., 2021; Wu et al., 2021a; Baden et al., 2021; Russell et al., 2021). Nowadays, the global vaccination initiatives cover 56% of the world population with two doses. Global vaccination success was limited by inaccessibility to vaccines and refusal to be vaccinated for personal reasons. In addition, significant efforts were placed in the study of repurposing drugs that may limit mortality and ameliorate COVID-19 symptoms (Wang et al., 2020b; Canedo-Marroquín et al., 2020; Valle et al., 2020). There are more than 2000 clinical trials registered on www.clinicaltrials.gov with various topics ranging from contact tracing, dietary supplements, anti-viral therapies, and drugs that have been in the spotlight, such as chloroquine, hydroxychloroquine, and ivermectin.
The current development of antibodies against SARS-CoV-2 focuses on neutralizing antibodies against the spike protein (Chi et al., 2020a; Shi et al., 2020). In the early days of the first wave of the COVID-19 outbreak, plasmapheresis of convalescent SARS-CoV-2 patients was implemented to supplement antibodies to those at risk due to the lack of alternatives (Jiang et al., 2020; Liu et al., 2020; Longueira et al., 2021) (Table 1).
The first neutralizing nanobodies against SARS-CoV-2 targeted SARS-CoV-1 and MERS-CoV RBDs, but fortunately, they also exhibited a remarkable cross-reactivity and neutralization capability against SARS-CoV-2 (Wrapp et al., 2020). These were followed by nanobodies identified from synthetic libraries, “sybodies”, against SARS-CoV-2 (Dong et al., 2020a; Custódio et al., 2020; Walter et al., 2020). The framework regions of the synthetic library were partially humanized to decrease the immune response if administered to humans.
Other reported strategies were the generation of a platform to develop SARS-CoV-2-specific single-domain antibodies of human origin (Wu et al., 2020), humanizing the nanobody backbone, and reducing potential immune recognition. Other studies reported successful isolation of nanobodies and their fusion to the human IgG1-Fc region, improving their binding and neutralizing capabilities (Chi et al., 2020b; Xiaojie et al., 2020; Lu et al., 2021a; Valenzuela Nieto et al., 2021) (Figure 1C).
Also, another nanobody set was isolated by from yeast surface-displayed synthetic library against epitopes of the SARS-CoV-2 spike protein. The researchers used Nb6 to design bivalent and trivalent nanobodies resulting in a 2000-fold increase in inhibitory activity against both pseudo virus and live SARS-CoV-2 in infection assays (Schoof et al., 2020) (Figure 1D).
Another group reported the isolation and characterization of an alpaca-derived single domain antibody fragment, Ty1, against spike protein (Hanke et al., 2020a; Hanke et al., 2020b). In this study, Ty1 was fused to an Fc domain, increasing the neutralizing capabilities of the nanobody. Moreover, in contrast to Nb6 that binds RBD in the down conformation (Figure 1D), cryo-electron microscopy studies demonstrated that Ty1 binds to an epitope on the RBD, accessible in both the “up” and “down” conformations (Figure 1E). Glycosylation sites N165, N234, and N343 on the spike protein shield RBD from conventional antibodies, especially when the RBD is in a “down” conformation (Watanabe et al., 2020). In the RBD-down conformation, the glycan on N165 points towards the Ty1-binding epitope, likely not leaving sufficient space to accommodate a conventional antibody. This indicates that nanobodies most likely recognize more epitopes for SARS-CoV-2 neutralization than conventional antibodies. Interestingly, a group of multivalent nanobodies bind the RBD domain of spike and lock it in the “up” conformation, a state that is typically associated with receptor binding activation. The premature activation of the fusion machinery on virions enhances neutralization in a non-reversible manner (Koenig et al., 2021).
Not only alpacas and llamas have been contributing for the generation of SARS-CoV-2 neutralizing nanobodies, also nanobodies isolated from a semisynthetic shark-derived library have been shown to neutralize SARS-CoV-2 (Gauhar et al., 2021).
Discussion
During the COVID-19 pandemic, new technologies were developed for the rapid isolation of nanobodies. Our team implemented a new procedure for fast, economical, and efficient selection of high affinity nanobodies based on bacterial display and density gradient (Valenzuela Nieto et al., 2021).
Unfortunately, new SARS-CoV-2 variants significantly scape the immune response raised by either vaccination or previous SARS-CoV-2 infections. The most dramatic example are the Omicron variants. Omicron BA1 has become the infectious virus in the history of humanity. Several mutations on the spike protein generated less efficient cleavage of the S1 domain by the furin protease TMPRSS2 at the cell membrane. Consequently, the infection predominantly affects the upper airway but is less severe in human and animal models (Chen et al., 2022; Wrenn et al., 2022). However, due to the substantial number of infected people, death rates increased worldwide. Furthermore, a second subvariant of Omicron, BA2, developed in parallel. BA2 preserves the immune evasion capabilities of BA1, but unfortunately, current reports indicate a higher lethality (Wolter et al., 2022). The mechanisms behind the high severity of Omicron BA2 infection are not well understood.
More than 2 years after the first COVID-19 outbreak, over six million succumbed to the disease. There were four infective waves worldwide, and currently, infection rates are rising again, suggesting we are entering another wave of COVID-19 caused by Omicron subvariants. Vaccines have been beneficial and saved millions of lives; however, we must take new complementary approaches due to immune escape.
Our immune responses as humans are determined by the way our immune system is organized, for instance, we cannot raise single chain antibodies as part of our antibody defense. The pandemic has challenged the human population with a virus that replicates, mutates, evades, and overcomes our immune response. Nanobodies differ from our own defense and provide new possibilities for the generation of effective neutralizing antibodies that we could never develop ourselves.
Author Contributions
Conceptualization: GV-N and AR-F. All authors have contributed to writing, reviewing, and editing sections and have agreed to the published version of the manuscript.
Funding
This work was funded by FONDECYT, No. 1200427; the regional Council of the “Los Rios region” projects FICR19-20 and FICR21-01; the Bio and Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (NRF-2020M3A9H5112394) to AR-F; postdoctoral fellowship ANID 3220635 to GV-N; graduate fellowship ANID N° 21161365 to RJ; ANID N°21160481 to A.C; and ANID N° 22170632 to CS; ZM-C was funded by the academic graduate student fellowship of the University of Costa Rica of the academic mobility program, and AB was funded by CORFO Project 19 IRLR–110191.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelghany, M. T., and Baggett, M. V. (2016). Caplacizumab for Acquired Thrombotic Thrombocytopenic Purpura. N. Engl. J. Med. 374 (25), 2497–2498. doi:10.1056/NEJMc1603180
Ahmad, J., Jiang, J., Boyd, L. F., Zeher, A., Huang, R., Xia, D., et al. (2021). Structures of Synthetic Nanobody-SARS-CoV-2 Receptor-Binding Domain Complexes Reveal Distinct Sites of Interaction. J. Biol. Chem. 297 (4), 101202. doi:10.1016/j.jbc.2021.101202
Anderson, G., Shriver-Lake, L., Walper, S., Ashford, L., Zabetakis, D., Liu, J., et al. (2018). Genetic Fusion of an Anti-BclA Single-Domain Antibody with Beta Galactosidase. Antibodies 7 (4), 36. doi:10.3390/antib7040036
Arbabi Ghahroudi, M., Desmyter, A., Wyns, L., Hamers, R., and Muyldermans, S. (1997). Selection and Identification of Single Domain Antibody Fragments from Camel Heavy-Chain Antibodies. FEBS Lett. 414 (3), 521–526. doi:10.1016/s0014-5793(97)01062-4
Arbabi-Ghahroudi, M. (2017). Camelid Single-Domain Antibodies: Historical Perspective and Future Outlook. Front. Immunol. 8, 1589. doi:10.3389/fimmu.2017.01589
Arbabi-Ghahroudi, M., Tanha, J., and MacKenzie, R. (2005). Prokaryotic Expression of Antibodies. Cancer Metastasis Rev. 24 (4), 501–519. doi:10.1007/s10555-005-6193-1
Ashour, J., Schmidt, F. I., Hanke, L., Cragnolini, J., Cavallari, M., Altenburg, A., et al. (2015). Intracellular Expression of Camelid Single-Domain Antibodies Specific for Influenza Virus Nucleoprotein Uncovers Distinct Features of its Nuclear Localization. J. Virol. 89 (5), 2792–2800. doi:10.1128/JVI.02693-14
Awadasseid, A., Wu, Y., Tanaka, Y., and Zhang, W. (2021). Current Advances in the Development of SARS-CoV-2 Vaccines. Int. J. Biol. Sci. 17 (1), 8–19. doi:10.7150/ijbs.52569
Baden, L. R., El Sahly, H. M., Essink, B., Kotloff, K., Frey, S., Novak, R., et al. (2021). Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 384 (5), 403–416. doi:10.1056/NEJMoa2035389
Bao, K., Liu, X., Xu, Q., Su, B., Liu, Z., Cao, H., et al. (2021). Nanobody Multimerization Strategy to Enhance the Sensitivity of Competitive ELISA for Detection of Ochratoxin A in Coffee Samples. Food control. 127, 108167. doi:10.1016/j.foodcont.2021.108167
Billen, B., Vincke, C., Hansen, R., Devoogdt, N., Muyldermans, S., Adriaensens, P., et al. (2017). Cytoplasmic versus Periplasmic Expression of Site-Specifically and Bioorthogonally Functionalized Nanobodies Using Expressed Protein Ligation. Protein Expr. Purif. 133, 25–34. doi:10.1016/j.pep.2017.02.009
Bond, C. J., Marsters, J. C., and Sidhu, S. S. (2003). Contributions of CDR3 to VHH Domain Stability and the Design of Monobody Scaffolds for Naive Antibody Libraries. J. Mol. Biol. 332 (3), 643–655. doi:10.1016/s0022-2836(03)00967-7
Camacho-Villegas, T., Mata-Gonzalez, T., Paniagua-Solis, J., Sanchez, E., and Licea, A. (2013). Human TNF Cytokine Neutralization with a vNAR fromHeterodontus Franciscishark: A Potential Therapeutic Use. MAbs 5 (1), 80–85. doi:10.4161/mabs.22593
Canedo-Marroquín, G., Saavedra, F., Andrade, C. A., Berrios, R. V., Rodríguez-Guilarte, L., Opazo, M. C., et al. (2020). SARS-CoV-2: Immune Response Elicited by Infection and Development of Vaccines and Treatments. Front. Immunol. 11, 569760. doi:10.3389/fimmu.2020.569760
Chakravarty, R., Goel, S., and Cai, W. (2014). Nanobody: the "magic Bullet" for Molecular Imaging? Theranostics 4 (4), 386–398. doi:10.7150/thno.8006
Chen, F., Liu, Z., and Jiang, F. (2021). Prospects of Neutralizing Nanobodies against SARS-CoV-2. Front. Immunol. 12, 690742. doi:10.3389/fimmu.2021.690742
Chen, J., Wang, R., Gilby, N. B., and Wei, G.-W. (2022). Omicron Variant (B.1.1.529): Infectivity, Vaccine Breakthrough, and Antibody Resistance. J. Chem. Inf. Model. 62 (2), 412–422. doi:10.1021/acs.jcim.1c01451
Chen, W., Zhu, Z., Feng, Y., and Dimitrov, D. S. (2008). Human Domain Antibodies to Conserved Sterically Restricted Regions on Gp120 as Exceptionally Potent Cross-Reactive HIV-1 Neutralizers. Proc. Natl. Acad. Sci. U.S.A. 105 (44), 17121–17126. doi:10.1073/pnas.0805297105
Cheong, W. S., Leow, C. Y., Abdul Majeed, A. B., and Leow, C. H. (2020). Diagnostic and Therapeutic Potential of Shark Variable New Antigen Receptor (VNAR) Single Domain Antibody. Int. J. Biol. Macromol. 147, 369–375. doi:10.1016/j.ijbiomac.2020.01.039
Chi, X., Liu, X., Wang, C., Zhang, X., Li, X., Hou, J., et al. (2020). Humanized Single Domain Antibodies Neutralize SARS-CoV-2 by Targeting the Spike Receptor Binding Domain. Nat. Commun. 11 (1), 4528. doi:10.1038/s41467-020-18387-8
Chi, X., Yan, R., Zhang, J., Zhang, G., Zhang, Y., Hao, M., et al. (2020). A Neutralizing Human Antibody Binds to the N-Terminal Domain of the Spike Protein of SARS-CoV-2. Science 369 (6504), 650–655. doi:10.1126/science.abc6952
Chi, X., Zhang, X., Pan, S., Yu, Y., Shi, Y., Lin, T., et al. (2022). An Ultrapotent RBD-Targeted Biparatopic Nanobody Neutralizes Broad SARS-CoV-2 Variants. Sig Transduct. Target Ther. 7 (1), 44. doi:10.1038/s41392-022-00912-4
Conrath, K. E., Lauwereys, M., Galleni, M., Matagne, A., Frere, J.-M., Kinne, J., et al. (2001). β-Lactamase Inhibitors Derived from Single-Domain Antibody Fragments Elicited in the Camelidae. Antimicrob. Agents Chemother. 45 (10), 2807–2812. doi:10.1128/AAC.45.10.2807-2812.2001
Conroy, P. J., Hearty, S., Leonard, P., and O’Kennedy, R. J. (2009). Antibody Production, Design and Use for Biosensor-Based Applications. Seminars Cell & Dev. Biol. 20 (1), 10–26. doi:10.1016/j.semcdb.2009.01.010
Custódio, T. F., Das, H., Sheward, D. J., Hanke, L., Pazicky, S., Pieprzyk, J., et al. (2020). Selection, Biophysical and Structural Analysis of Synthetic Nanobodies that Effectively Neutralize SARS-CoV-2. Nat. Commun. 11 (1), 5588. doi:10.1038/s41467-020-19204-y
de Marco, A. (2020). Recombinant Expression of Nanobodies and Nanobody-Derived Immunoreagents. Protein Expr. Purif. 172, 105645. doi:10.1016/j.pep.2020.105645
De Munter, S., Ingels, J., Goetgeluk, G., Bonte, S., Pille, M., Weening, K., et al. (2018). Nanobody Based Dual Specific Cars. Ijms 19 (2), 403. doi:10.3390/ijms19020403
Debnath, S., Zhou, N., McLaughlin, M., Rice, S., Pillai, A. K., Hao, G., et al. (2022). PSMA-targeting Imaging and Theranostic Agents-Current Status and Future Perspective. Ijms 23 (3), 1158. doi:10.3390/ijms23031158
Deckers, N., Saerens, D., Kanobana, K., Conrath, K., Victor, B., Wernery, U., et al. (2009). Nanobodies, a Promising Tool for Species-specific Diagnosis of Taenia Solium Cysticercosis. Int. J. Parasitol. 39 (5), 625–633. doi:10.1016/j.ijpara.2008.10.012
Desmyter, A., Transue, T. R., Ghahroudi, M. A., Dao Thi, M.-H., Poortmans, F., Hamers, R., et al. (1996). Crystal Structure of a Camel Single-Domain VH Antibody Fragment in Complex with Lysozyme. Nat. Struct. Mol. Biol. 3 (9), 803–811. doi:10.1038/nsb0996-803
Dong, J., Huang, B., Jia, Z., Wang, B., Gallolu Kankanamalage, S., Titong, A., et al. (2020). Development of Multi-specific Humanized Llama Antibodies Blocking SARS-CoV-2/ace2 Interaction with High Affinity and Avidity. Emerg. microbes Infect. 9 (1), 1034–1036. doi:10.1080/22221751.2020.1768806
Dong, J., Huang, B., Wang, B., Titong, A., Gallolu Kankanamalage, S., Jia, Z., et al. (2020). Development of Humanized Tri-specific Nanobodies with Potent Neutralization for SARS-CoV-2. Sci. Rep. 10 (1), 17806. doi:10.1038/s41598-020-74761-y
Dooley, H., and Flajnik, M. F. (2006). Antibody Repertoire Development in Cartilaginous Fish. Dev. Comp. Immunol. 30 (1-2), 43–56. doi:10.1016/j.dci.2005.06.022
Dooley, H., Stanfield, R. L., Brady, R. A., and Flajnik, M. F. (2006). First Molecular and Biochemical Analysis of In Vivo Affinity Maturation in an Ectothermic Vertebrate. Proc. Natl. Acad. Sci. U.S.A. 103 (6), 1846–1851. doi:10.1073/pnas.0508341103
Dörner, T., Weinblatt, M., and Beneden, K. V. (2017). “FRI0239 Results of a Phase 2b Study of Vobarilizumab, an Anti-interleukin-6 Receptor Nanobody, as Monotherapy in Patients with Moderate to Severe Rheumatoid Arthritis,” in Poster Presentations (BMJ Publishing Group Ltd and European League Against Rheumatism), 575, 2–575.
Du, T., Zhu, G., Wu, X., Fang, J., and Zhou, E.-M. (2019). Biotinylated Single-Domain Antibody-Based Blocking ELISA for Detection of Antibodies against Swine Influenza Virus. Ijn 14, 9337–9349. doi:10.2147/IJN.S218458
Esparza, T. J., Martin, N. P., Anderson, G. P., Goldman, E. R., and Brody, D. L. (2020). High Affinity Nanobodies Block SARS-CoV-2 Spike Receptor Binding Domain Interaction with Human Angiotensin Converting Enzyme. Sci. Rep. 10 (1), 22370. doi:10.1038/s41598-020-79036-0
Esposito, G., Hunashal, Y., Percipalle, M., Venit, T., Dieng, M. M., Fogolari, F., et al. (2021). NMR‐Based Analysis of Nanobodies to SARS‐CoV‐2 Nsp9 Reveals a Possible Antiviral Strategy against COVID‐19. Adv. Biol. 5 (12), 2101113. doi:10.1002/adbi.202101113
Fatima, A., Wang, H., Kang, K., Xia, L., Wang, Y., Ye, W., et al. (2014). Development of VHH Antibodies against Dengue Virus Type 2 NS1 and Comparison with Monoclonal Antibodies for Use in Immunological Diagnosis. Plos One 9 (4), e95263. doi:10.1371/journal.pone.0095263
Favorskaya, I. A., Shcheblyakov, D. V., Esmagambetov, I. B., Dolzhikova, I. V., Alekseeva, I. A., Korobkova, A. I., et al. (2022). Single-Domain Antibodies Efficiently Neutralize SARS-CoV-2 Variants of Concern. Front. Immunol. 13, 822159. doi:10.3389/fimmu.2022.822159
Feige, M. J., Gräwert, M. A., Marcinowski, M., Hennig, J., Behnke, J., Ausländer, D., et al. (2014). The Structural Analysis of Shark IgNAR Antibodies Reveals Evolutionary Principles of Immunoglobulins. Proc. Natl. Acad. Sci. U.S.A. 111 (22), 8155–8160. doi:10.1073/pnas.1321502111
Frenken, L. G. J., van der Linden, R. H. J., Hermans, P. W. J. J., Bos, J. W., Ruuls, R. C., de Geus, B., et al. (2000). Isolation of Antigen Specific Llama VHH Antibody Fragments and Their High Level Secretion by Saccharomyces cerevisiae. J. Biotechnol. 78 (1), 11–21. doi:10.1016/S0168-1656(99)00228-X
Frigerio, B., Luison, E., Desideri, A., Iacovelli, F., Camisaschi, C., Seregni, E. C., et al. (2021). Validity of Anti-PSMA ScFvD2B as a Theranostic Tool: A Narrative-Focused Review. Biomedicines 9 (12), 1870. doi:10.3390/biomedicines9121870
Gai, J., Ma, L., Li, G., Zhu, M., Qiao, P., Li, X., et al. (2021). A Potent Neutralizing Nanobody against SARS‐CoV‐2 with Inhaled Delivery Potential. MedComm 2 (1), 101–113. doi:10.1002/mco2.60
Gainkam, L. O. T., Huang, L., Caveliers, V., Keyaerts, M., Hernot, S., Vaneycken, I., et al. (2008). Comparison of the Biodistribution and Tumor Targeting of Two 99mTc-Labeled Anti-EGFR Nanobodies in Mice, Using Pinhole SPECT/micro-CT. J. Nucl. Med. 49 (5), 788–795. doi:10.2967/jnumed.107.048538
Gauhar, A., Privezentzev, C. V., Demydchuk, M., Gerlza, T., Rieger, J., Kungl, A. J., et al. (2021). Single Domain Shark VNAR Antibodies Neutralize SARS‐CoV‐2 Infection In Vitro. FASEB J. 35 (11), e21970. doi:10.1096/fj.202100986RR
Girt, G. C., Lakshminarayanan, A., Huo, J., Dormon, J., Norman, C., Afrough, B., et al. (2021). The Use of Nanobodies in a Sensitive ELISA Test for SARS-CoV-2 Spike 1 Protein. R. Soc. open Sci. 8 (9), 211016. doi:10.1098/rsos.211016
Gong, R., Wang, Y., Ying, T., and Dimitrov, D. S. (2012). Bispecific Engineered Antibody Domains (Nanoantibodies) that Interact Noncompetitively with an HIV-1 Neutralizing Epitope and FcRn. Plos One 7 (8), e42288. doi:10.1371/journal.pone.0042288
Goossens, J., Sein, H., Lu, S., Radwanska, M., Muyldermans, S., Sterckx, Y. G.-J., et al. (2017). Functionalization of Gold Nanoparticles with Nanobodies through Physical Adsorption. Anal. Methods 9 (23), 3430–3440. doi:10.1039/C7AY00854F
Götzke, H., Kilisch, M., Martínez-Carranza, M., Sograte-Idrissi, S., Rajavel, A., Schlichthaerle, T., et al. (2019). The ALFA-Tag Is a Highly Versatile Tool for Nanobody-Based Bioscience Applications. Nat. Commun. 10 (1), 4403. doi:10.1038/s41467-019-12301-7
Govaert, J., Pellis, M., Deschacht, N., Vincke, C., Conrath, K., Muyldermans, S., et al. (2012). Dual Beneficial Effect of Interloop Disulfide Bond for Single Domain Antibody Fragments. J. Biol. Chem. 287 (3), 1970–1979. doi:10.1074/jbc.M111.242818
Gray, E. R., Brookes, J. C., Caillat, C., Turbé, V., Webb, B. L. J., Granger, L. A., et al. (2017). Unravelling the Molecular Basis of High Affinity Nanobodies against HIV P24: In Vitro Functional, Structural, and In Silico Insights. ACS Infect. Dis. 3 (7), 479–491. doi:10.1021/acsinfecdis.6b00189
Günaydın, G., Álvarez, B., Lin, Y., Hammarström, L., and Marcotte, H. (2014). Co-expression of Anti-rotavirus Proteins (Llama VHH Antibody Fragments) in Lactobacillus: Development and Functionality of Vectors Containing Two Expression Cassettes in Tandem. Plos One 9 (4), e96409. doi:10.1371/journal.pone.0096409
Guo, K., Wustoni, S., Koklu, A., Díaz-Galicia, E., Moser, M., Hama, A., et al. (2021). Rapid Single-Molecule Detection of COVID-19 and MERS Antigens via Nanobody-Functionalized Organic Electrochemical Transistors. Nat. Biomed. Eng. 5 (7), 666–677. doi:10.1038/s41551-021-00734-9
Güttler, T., Aksu, M., Dickmanns, A., Stegmann, K. M., Gregor, K., Rees, R., et al. (2021). Neutralization of SARS‐CoV‐2 by Highly Potent, Hyperthermostable, and Mutation‐tolerant Nanobodies. Embo J. 40 (19), e107985. doi:10.15252/embj.2021107985
Haga, K., Takai-Todaka, R., Matsumura, Y., Song, C., Takano, T., Tojo, T., et al. (2021). Nasal Delivery of Single-Domain Antibody Improves Symptoms of SARS-CoV-2 Infection in an Animal Model. PLoS Pathog. 17 (10), e1009542. doi:10.1371/journal.ppat.1009542
Hagihara, Y., and Saerens, D. (2014). Engineering Disulfide Bonds within an Antibody. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1844 (11), 2016–2023. doi:10.1016/j.bbapap.2014.07.005
Hamers-Casterman, C., Atarhouch, T., Muyldermans, S., Robinson, G., Hammers, C., Songa, E. B., et al. (1993). Naturally Occurring Antibodies Devoid of Light Chains. Nature 363 (6428), 446–448. doi:10.1038/363446a0
Hanke, L., Das, H., Sheward, D. J., Perez Vidakovics, L., Urgard, E., Moliner-Morro, A., et al. (2022). A Bispecific Monomeric Nanobody Induces Spike Trimer Dimers and Neutralizes SARS-CoV-2 In Vivo. Nat. Commun. 13 (1), 155. doi:10.1038/s41467-021-27610-z
Hanke, L., Perez, L. V., Sheward, D. J., Das, H., Schulte, T., Moliner-Morro, A., et al. (2020). An Alpaca Nanobody Neutralizes SARS-CoV-2 by Blocking Receptor Interaction. BioRxiv. doi:10.1101/2020.06.02.130161
Hanke, L., Vidakovics Perez, L., Sheward, D. J., Das, H., Schulte, T., Moliner-Morro, A., et al. (2020). An Alpaca Nanobody Neutralizes SARS-CoV-2 by Blocking Receptor Interaction. Nat. Commun. 11 (1), 4420. doi:10.1038/s41467-020-18174-5
Harmand, T. J., Islam, A., Pishesha, N., and Ploegh, H. L. (2021). Nanobodies as In Vivo, Non-invasive, Imaging Agents. RSC Chem. Biol. 2 (3), 685–701. doi:10.1039/d1cb00023c
Hassanzadeh Gh, G., De Silva, K. S., Dambly-Chaudière, C., Brys, L., Ghysen, A., Hamers, R., et al. (1998). Isolation and Characterization of Single-Chain Fv Genes Encoding Antibodies Specific for Drosophila Poxn Protein. FEBS Lett. 437 (1-2), 75–80. doi:10.1016/s0014-5793(98)01204-6
Hernot, S., Unnikrishnan, S., Du, Z., Shevchenko, T., Cosyns, B., Broisat, A., et al. (2012). Nanobody-coupled Microbubbles as Novel Molecular Tracer. J. Control. Release 158 (2), 346–353. doi:10.1016/j.jconrel.2011.12.007
Huang, L., Muyldermans, S., and Saerens, D. (2010). Nanobodies: Proficient Tools in Diagnostics. Expert Rev. Mol. Diagnostics 10 (6), 777–785. doi:10.1586/erm.10.62
Huang, L., Reekmans, G., Saerens, D., Friedt, J.-M., Frederix, F., Francis, L., et al. (2005). Prostate-specific Antigen Immunosensing Based on Mixed Self-Assembled Monolayers, Camel Antibodies and Colloidal Gold Enhanced Sandwich Assays. Biosens. Bioelectron. 21 (3), 483–490. doi:10.1016/j.bios.2004.11.016
Hultberg, A., Temperton, N. J., Rosseels, V., Koenders, M., Gonzalez-Pajuelo, M., Schepens, B., et al. (2011). Llama-derived Single Domain Antibodies to Build Multivalent, Superpotent and Broadened Neutralizing Anti-viral Molecules. Plos One 6 (4), e17665. doi:10.1371/journal.pone.0017665
Huo, J., Le Bas, A., Ruza, R. R., Duyvesteyn, H. M. E., Mikolajek, H., Malinauskas, T., et al. (2020). Neutralizing Nanobodies Bind SARS-CoV-2 Spike RBD and Block Interaction with ACE2. Nat. Struct. Mol. Biol. 27 (9), 846–854. doi:10.1038/s41594-020-0469-6
Huo, J., Mikolajek, H., Le Bas, A., Clark, J. J., Sharma, P., Kipar, A., et al. (2021). A Potent SARS-CoV-2 Neutralising Nanobody Shows Therapeutic Efficacy in the Syrian Golden Hamster Model of COVID-19. Nat. Commun. 12 (1), 5469. doi:10.1038/s41467-021-25480-z
Ismaili, A., Jalali Javaran, M., Rasaee, M. J., Rahbarizadeh, F., and Rajabi Memari, H. (2006). Cloning and Expression of Recombinant Camelid Single-Domain Antibody in Tobacco. Iran. J. Biotechnol.
Jiang, S., Hillyer, C., and Du, L. (2020). Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 41 (5), 355–359. doi:10.1016/j.it.2020.03.007
Jovčevska, I., and Muyldermans, S. (2020). The Therapeutic Potential of Nanobodies. BioDrugs 34 (1), 11–26. doi:10.1007/s40259-019-00392-z
Keen, N. T., and Tamaki, S. (1986). Structure of Two Pectate Lyase Genes from Erwinia Chrysanthemi EC16 and Their High-Level Expression in Escherichia coli. J. Bacteriol. 168 (2), 595–606. doi:10.1128/jb.168.2.595-606.1986
Kilisch, M., Götzke, H., Gere-Becker, M., Crauel, A., Opazo, F., and Frey, S. (2021). Discovery and Characterization of an ALFA-tag-specific Affinity Resin Optimized for Protein Purification at Low Temperatures in Physiological Buffer. Biomolecules 11 (2), 269. doi:10.3390/biom11020269
Koenig, P.-A., Das, H., Liu, H., Kümmerer, B. M., Gohr, F. N., Jenster, L.-M., et al. (2021). Structure-guided Multivalent Nanobodies Block SARS-CoV-2 Infection and Suppress Mutational Escape. Science 371, 6530. doi:10.1126/science.abe6230
Koromyslova, A. D., and Hansman, G. S. (2017). Nanobodies Targeting Norovirus Capsid Reveal Functional Epitopes and Potential Mechanisms of Neutralization. PLoS Pathog. 13 (11), e1006636. doi:10.1371/journal.ppat.1006636
Krainer, F. W., Gmeiner, C., Neutsch, L., Windwarder, M., Pletzenauer, R., Herwig, C., et al. (2013). Knockout of an Endogenous Mannosyltransferase Increases the Homogeneity of Glycoproteins Produced in Pichia pastoris. Sci. Rep. 3, 3279. doi:10.1038/srep03279
Langley, R. G., Elewski, B. E., Lebwohl, M., Reich, K., Griffiths, C. E. M., Papp, K., et al. (2014). Secukinumab in Plaque Psoriasis - Results of Two Phase 3 Trials. N. Engl. J. Med. 371 (4), 326–338. doi:10.1056/NEJMoa1314258
Larios Mora, A., and Gallup, L. (2018). Delivery of ALX-0171 by Inhalation Greatly Reduces Respiratory Syncytial Virus Disease in Newborn Lambs. MAbs 10 (5), 778–795. doi:10.1080/19420862.2018.1470727
Lauwereys, M., Arbabi Ghahroudi, M., Desmyter, A., et al. (1998). Potent Enzyme Inhibitors Derived from Dromedary Heavy-Chain Antibodies. EMBO J. 17 (13), 3512–3520. doi:10.1093/emboj/17.13.3512
Li, D., Ji, F., Huang, C., and Jia, L. (2019). High Expression Achievement of Active and Robust Anti-β2 Microglobulin Nanobodies via E.Coli Hosts Selection. Molecules 24 (16), 2860. doi:10.3390/molecules24162860
Li, J.-F., He, L., Deng, Y.-Q., Qi, S.-H., Chen, Y.-H., Zhang, X.-L., et al. (2021). Generation and Characterization of a Nanobody against SARS-CoV. Virol. Sin. 36 (6), 1484–1491. doi:10.1007/s12250-021-00436-1
Li, L., Guo, P., Zhang, X., Yu, Z., Zhang, W., and Sun, H. (2021). SARS-CoV-2 Vaccine Candidates in Rapid Development. Hum. vaccines Immunother. 17 (3), 644–653. doi:10.1080/21645515.2020.1804777
Li, T., Cai, H., Yao, H., Zhou, B., Zhang, N., van Vlissingen, M. F., et al. (2021). A Synthetic Nanobody Targeting RBD Protects Hamsters from SARS-CoV-2 Infection. Nat. Commun. 12 (1), 4635. doi:10.1038/s41467-021-24905-z
Li, Y.-D., Chi, W.-Y., Su, J.-H., Ferrall, L., Hung, C.-F., and Wu, T.-C. (2020). Coronavirus Vaccine Development: from SARS and MERS to COVID-19. J. Biomed. Sci. 27 (1), 104. doi:10.1186/s12929-020-00695-2
Liu, S. T. H., Lin, H.-M., Baine, I., Wajnberg, A., Gumprecht, J. P., Rahman, F., et al. (2020). Convalescent Plasma Treatment of Severe COVID-19: a Propensity Score-Matched Control Study. Nat. Med. 26 (11), 1708–1713. doi:10.1038/s41591-020-1088-9
Liu, Y., and Huang, H. (2018). Expression of Single-Domain Antibody in Different Systems. Appl. Microbiol. Biotechnol. 102 (2), 539–551. doi:10.1007/s00253-017-8644-3
Longueira, Y., Polo, M. L., Turk, G., Laufer, N., and Laufer, N. (2021). Dynamics of SARS-CoV-2-specific Antibodies Among COVID19 Biobank Donors in Argentina. Heliyon 7 (10), e08140. doi:10.1016/j.heliyon.2021.e08140
Lu, Q., Liu, J., Zhao, S., Gomez Castro, M. F., Laurent-Rolle, M., Dong, J., et al. (2021). SARS-CoV-2 Exacerbates Proinflammatory Responses in Myeloid Cells through C-type Lectin Receptors and Tweety Family Member 2. Immunity 54 (6), 1304–1319. e9. doi:10.1016/j.immuni.2021.05.006
Lu, Q., Zhang, Z., Li, H., Zhong, K., Zhao, Q., Wang, Z., et al. (2021). Development of Multivalent Nanobodies Blocking SARS-CoV-2 Infection by Targeting RBD of Spike Protein. J. Nanobiotechnol 19 (1), 33. doi:10.1186/s12951-021-00768-w
Ma, H., Zeng, W., Meng, X., Huang, X., Yang, Y., Zhao, D., et al. (2021). Potent Neutralization of SARS-CoV-2 by Hetero-Bivalent Alpaca Nanobodies Targeting the Spike Receptor-Binding Domain. J. Virol. 95. doi:10.1128/JVI.02438-20
Maggi, M., and Scotti, C. (2017). Enhanced Expression and Purification of Camelid Single Domain VHH Antibodies from Classical Inclusion Bodies. Protein Expr. Purif. 136, 39–44. doi:10.1016/j.pep.2017.02.007
Massoud, T. F., and Gambhir, S. S. (2003). Molecular Imaging in Living Subjects: Seeing Fundamental Biological Processes in a New Light. Genes Dev. 17 (5), 545–580. doi:10.1101/gad.1047403
Mast, F. D., Fridy, P. C., Ketaren, N. E., Wang, J., Jacobs, E. Y., Olivier, J. P., et al. (2021). Highly Synergistic Combinations of Nanobodies that Target SARS-CoV-2 and Are Resistant to Escape. eLife 10, 73027. doi:10.7554/eLife.73027
Matz, J., Kessler, P., Bouchet, J., Combes, O., Ramos, O. H. P., Barin, F., et al. (2013). Straightforward Selection of Broadly Neutralizing Single-Domain Antibodies Targeting the Conserved CD4 and Coreceptor Binding Sites of HIV-1 Gp120. J. Virol. 87 (2), 1137–1149. doi:10.1128/JVI.00461-12
McCoy, L. E., Quigley, A. F., Strokappe, N. M., Bulmer-Thomas, B., Seaman, M. S., Mortier, D., et al. (2012). Potent and Broad Neutralization of HIV-1 by a Llama Antibody Elicited by Immunization. J. Exp. Med. 209 (6), 1091–1103. doi:10.1084/jem.20112655
Muyldermans, S., Atarhouch, T., Saldanha, J., Barbosa, J. A. R. G., and Hamers, R. (1994). Sequence and Structure of VH Domain from Naturally Occurring Camel Heavy Chain Immunoglobulins Lacking Light Chains. Protein Eng. Des. Sel. 7 (9), 1129–1135. doi:10.1093/protein/7.9.1129
Muyldermans, S. (2013). Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 82, 775–797. doi:10.1146/annurev-biochem-063011-092449
Nambulli, S., Xiang, Y., Tilston-Lunel, N. L., Rennick, L. J., Sang, Z., Klimstra, W. B., et al. (2021). Inhalable Nanobody (PiN-21) Prevents and Treats SARS-CoV-2 Infections in Syrian Hamsters at Ultra-low Doses. Sci. Adv. 7 (22). doi:10.1126/sciadv.abh0319
Noor, A., Walser, G., Wesseling, M., Giron, P., Laffra, A.-M., Haddouchi, F., et al. (2018). Production of a Mono-Biotinylated EGFR Nanobody in the E. coli Periplasm Using the pET22b Vector. BMC Res. Notes 11 (1), 751. doi:10.1186/s13104-018-3852-1
Nuttall, S. D., Krishnan, U. V., Hattarki, M., De Gori, R., Irving, R. A., and Hudson, P. J. (2001). Isolation of the New Antigen Receptor from Wobbegong Sharks, and Use as a Scaffold for the Display of Protein Loop Libraries. Mol. Immunol. 38 (4), 313–326. doi:10.1016/s0161-5890(01)00057-8
Ohtani, M., Hikima, J.-i., Jung, T. S., Kondo, H., Hirono, I., and Aoki, T. (2013). Construction of an Artificially Randomized IgNAR Phage Display Library: Screening of Variable Regions that Bind to Hen Egg White Lysozyme. Mar. Biotechnol. 15 (1), 56–62. doi:10.1007/s10126-012-9456-1
Palomo, C., and Mas, V. (2016). Trivalency of a Nanobody Specific for the Human Respiratory Syncytial Virus Fusion Glycoprotein Drastically Enhances Virus Neutralization and Impacts Escape Mutant Selection. Antimicrob. Agents Chemother. 60 (11), 6498–6509. doi:10.1128/AAC.00842-16
Penault-Llorca, F., Cayre, A., Arnould, L., Bibeau, F., Bralet, M.-P., Rochaix, P., et al. (2006). Is There an Immunohistochemical Technique Definitively Valid in Epidermal Growth Factor Receptor Assessment? Oncol. Rep. 16 (6), 1173–1179. doi:10.3892/or.16.6.1173
Peyvandi, F., Scully, M., Kremer Hovinga, J. A., Cataland, S., Knöbl, P., Wu, H., et al. (2016). Caplacizumab for Acquired Thrombotic Thrombocytopenic Purpura. N. Engl. J. Med. 374 (6), 511–522. doi:10.1056/NEJMoa1505533
Peyvandi, F., Scully, M., Kremer Hovinga, J. A., Knöbl, P., Cataland, S., De Beuf, K., et al. (2017). Caplacizumab Reduces the Frequency of Major Thromboembolic Events, Exacerbations and Death in Patients with Acquired Thrombotic Thrombocytopenic Purpura. J. Thromb. Haemost. 15 (7), 1448–1452. doi:10.1111/jth.13716
Pinto Torres, J. E., Goossens, J., Ding, J., Li, Z., Lu, S., Vertommen, D., et al. (2018). Development of a Nanobody-Based Lateral Flow Assay to Detect Active Trypanosoma Congolense Infections. Sci. Rep. 8 (1), 9019. doi:10.1038/s41598-018-26732-7
Pymm, P., Adair, A., Chan, L.-J., Cooney, J. P., Mordant, F. L., Allison, C. C., et al. (2021). Nanobody Cocktails Potently Neutralize SARS-CoV-2 D614G N501Y Variant and Protect Mice. Proc. Natl. Acad. Sci. U.S.A. 118 (19), 118. doi:10.1073/pnas.2101918118
Qin, X., Duan, M., Pei, D., Lin, J., Wang, L., Zhou, P., et al. (2021). Development of Novel-Nanobody-Based Lateral-Flow Immunochromatographic Strip Test for Rapid Detection of Recombinant Human Interferon α2b. J. Pharm. Analysis 12, 308–316. doi:10.1016/j.jpha.2021.07.003
Rosano, G. L., Morales, E. S., and Ceccarelli, E. A. (2019). New Tools for Recombinant Protein Production in Escherichia coli : A 5‐year Update. Protein Sci. 28 (8), 1412–1422. doi:10.1002/pro.3668
Roux, K. H., Greenberg, A. S., Greene, L., Strelets, L., Avila, D., McKinney, E. C., et al. (1998). Structural Analysis of the Nurse Shark (New) Antigen Receptor (NAR): Molecular Convergence of NAR and Unusual Mammalian Immunoglobulins. Proc. Natl. Acad. Sci. U.S.A. 95 (20), 11804–11809. doi:10.1073/pnas.95.20.11804
Ruano-Gallego, D., Fraile, S., Gutierrez, C., and Fernández, L. Á. (2019). Screening and Purification of Nanobodies from E. coli Culture Supernatants Using the Hemolysin Secretion System. Microb. Cell Fact. 18 (1), 47. doi:10.1186/s12934-019-1094-0
Russell, R. L., Pelka, P., and Mark, B. L. (2021). Frontrunners in the Race to Develop a SARS-CoV-2 Vaccine. Can. J. Microbiol. 67 (3), 189–212. doi:10.1139/cjm-2020-0465
Saerens, D., Frederix, F., Reekmans, G., Conrath, K., Jans, K., Brys, L., et al. (2005). Engineering Camel Single-Domain Antibodies and Immobilization Chemistry for Human Prostate-specific Antigen Sensing. Anal. Chem. 77 (23), 7547–7555. doi:10.1021/ac051092j
Saerens, D., Huang, L., Bonroy, K., and Muyldermans, S. (2008). Antibody Fragments as Probe in Biosensor Development. Sensors 8 (8), 4669–4686. doi:10.3390/s8084669
Saerens, D., Stijlemans, B., Baral, T. N., Nguyen Thi, G. T., Wernery, U., Magez, S., et al. (2008). Parallel Selection of Multiple Anti-infectome Nanobodies without Access to Purified Antigens. J. Immunol. Methods 329 (1-2), 138–150. doi:10.1016/j.jim.2007.10.005
Salema, V., and Fernández, L. Á. (2013). High Yield Purification of Nanobodies from the Periplasm of E. coli as Fusions with the Maltose Binding Protein. Protein Expr. Purif. 91 (1), 42–48. doi:10.1016/j.pep.2013.07.001
Salema, V., Marín, E., Martínez-Arteaga, R., Ruano-Gallego, D., Fraile, S., Margolles, Y., et al. (2013). Selection of Single Domain Antibodies from Immune Libraries Displayed on the Surface of E. coli Cells with Two β-Domains of Opposite Topologies. PLoS One 8 (9), e75126. doi:10.1371/journal.pone.0075126
Sang, Z., Xiang, Y., Bahar, I., and Shi, Y. (2022). Llamanade: An Open-Source Computational Pipeline for Robust Nanobody Humanization. Structure 30 (3), 418–429. doi:10.1016/j.str.2021.11.006
Schepens, B., Ibañez, L. I., De Baets, S., Hultberg, A., Bogaert, P., De Bleser, P., et al. (2011). Nanobodies Specific for Respiratory Syncytial Virus Fusion Protein Protect against Infection by Inhibition of Fusion. J. Infect. Dis. 204 (11), 1692–1701. doi:10.1093/infdis/jir622
Schepens, B., van Schie, L., Nerinckx, W., Roose, K., Van Breedam, W., Fijalkowska, D., et al. (2021). An Affinity-Enhanced, Broadly Neutralizing Heavy Chain-Only Antibody Protects against SARS-CoV-2 Infection in Animal Models. Sci. Transl. Med. 13 (621), eabi7826. doi:10.1126/scitranslmed.abi7826
Schmidt, F. I., Hanke, L., Morin, B., Brewer, R., Brusic, V., Whelan, S. P. J., et al. (2016). Phenotypic Lentivirus Screens to Identify Functional Single Domain Antibodies. Nat. Microbiol. 1 (8), 16080. doi:10.1038/nmicrobiol.2016.80
Schoof, M., Faust, B., Saunders, R. A., Sangwan, S., Rezelj, V., Hoppe, N., et al. (2020). An Ultrapotent Synthetic Nanobody Neutralizes SARS-CoV-2 by Stabilizing Inactive Spike. Science 370 (6523), 1473–1479. doi:10.1126/science.abe3255
Scully, M., Cataland, S. R., Peyvandi, F., Coppo, P., Knöbl, P., Kremer Hovinga, J. A., et al. (2019). Caplacizumab Treatment for Acquired Thrombotic Thrombocytopenic Purpura. N. Engl. J. Med. 380 (4), 335–346. doi:10.1056/NEJMoa1806311
Sheng, Y., Wang, K., Lu, Q., Ji, P., Liu, B., Zhu, J., et al. (2019). Nanobody-horseradish Peroxidase Fusion Protein as an Ultrasensitive Probe to Detect Antibodies against Newcastle Disease Virus in the Immunoassay. J. Nanobiotechnol 17 (1), 35. doi:10.1186/s12951-019-0468-0
Shi, R., Shan, C., Duan, X., Chen, Z., Liu, P., Song, J., et al. (2020). A Human Neutralizing Antibody Targets the Receptor-Binding Site of SARS-CoV-2. Nature 584 (7819), 120–124. doi:10.1038/s41586-020-2381-y
Shi, Z., Li, X., Wang, L., Sun, Z., Zhang, H., Chen, X., et al. (2022). Structural Basis of Nanobodies Neutralizing SARS-CoV-2 Variants. Structure 30, 707–720. doi:10.1016/j.str.2022.02.011
Shriver-Lake, L. C., Goldman, E. R., Zabetakis, D., and Anderson, G. P. (2017). Improved Production of Single Domain Antibodies with Two Disulfide Bonds by Co-expression of Chaperone Proteins in the Escherichia coli Periplasm. J. Immunol. Methods 443, 64–67. doi:10.1016/j.jim.2017.01.007
Sograte-Idrissi, S., Schlichthaerle, T., Duque-Afonso, C. J., Alevra, M., Strauss, S., Moser, T., et al. (2020). Circumvention of Common Labelling Artefacts Using Secondary Nanobodies. Nanoscale 12 (18), 10226–10239. doi:10.1039/d0nr00227e
Stanfield, R. L., Dooley, H., Flajnik, M. F., and Wilson, I. A. (2004). Crystal Structure of a Shark Single-Domain Antibody V Region in Complex with Lysozyme. Science 305 (5691), 1770–1773. doi:10.1126/science.1101148
Stefan, M. A., Light, Y. K., Schwedler, J. L., McIlroy, P. R., Courtney, C. M., Saada, E. A., et al. (2021). Development of Potent and Effective Synthetic SARS-CoV-2 Neutralizing Nanobodies. MAbs 13 (1), 1958663. doi:10.1080/19420862.2021.1958663
Stijlemans, B., Conrath, K., Cortez-Retamozo, V., Van Xong, H., Wyns, L., Senter, P., et al. (2004). Efficient Targeting of Conserved Cryptic Epitopes of Infectious Agents by Single Domain Antibodies. J. Biol. Chem. 279 (2), 1256–1261. doi:10.1074/jbc.M307341200
Stijlemans, B., De Baetselier, P., Caljon, G., Van Den Abbeele, J., Van Ginderachter, J. A., and Magez, S. (2017). Nanobodies as Tools to Understand, Diagnose, and Treat African Trypanosomiasis. Front. Immunol. 8. doi:10.3389/fimmu.2017.00724
Stohr, L., and Palomo, T. (2016). Generation and Characterization of ALX-0171, a Potent Novel Therapeutic Nanobody for the Treatment of Respiratory Syncytial Virus Infection. Antimicrob. Agents Chemother. 60 (1), 6–13. doi:10.1128/AAC.01802-15
Sun, D., Sang, Z., Kim, Y. J., Xiang, Y., Cohen, T., Belford, A. K., et al. (2021). Potent Neutralizing Nanobodies Resist Convergent Circulating Variants of SARS-CoV-2 by Targeting Diverse and Conserved Epitopes. Nat. Commun. 12 (1), 4676. doi:10.1038/s41467-021-24963-3
Sun, Z., Lv, J., Liu, X., Tang, Z., Wang, X., Xu, Y., et al. (2018). Development of a Nanobody-AviTag Fusion Protein and its Application in a Streptavidin-Biotin-Amplified Enzyme-Linked Immunosorbent Assay for Ochratoxin A in Cereal. Anal. Chem. 90 (17), 10628–10634. doi:10.1021/acs.analchem.8b03085
Svecova, D., Lubell, M. W., Casset-Semanaz, F., Mackenzie, H., Grenningloh, R., and Krueger, J. G. (2019). A Randomized, Double-Blind, Placebo-Controlled Phase 1 Study of Multiple Ascending Doses of Subcutaneous M1095, an Anti-interleukin 17A/F Nanobody, in Moderate-To-Severe Psoriasis. J. Am. Acad. Dermatology 81 (1), 196–203. doi:10.1016/j.jaad.2019.03.056
Tarr, A. W., Lafaye, P., Meredith, L., Damier-Piolle, L., Urbanowicz, R. A., Meola, A., et al. (2013). An Alpaca Nanobody Inhibits Hepatitis C Virus Entry and Cell-To-Cell Transmission. Hepatology 58 (3), 932–939. doi:10.1002/hep.26430
Tijink, B. M., Laeremans, T., Budde, M., Walsum, M. S.-v., Dreier, T., de Haard, H. J., et al. (2008). Improved Tumor Targeting of Anti-epidermal Growth Factor Receptor Nanobodies through Albumin Binding: Taking Advantage of Modular Nanobody Technology. Mol. Cancer Ther. 7 (8), 2288–2297. doi:10.1158/1535-7163.MCT-07-2384
To, K. K.-W., Tsang, O. T.-Y., Yip, C. C.-Y., Chan, K.-H., Wu, T.-C., Chan, J. M.-C., et al. (2020). Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 71 (15), 841–843. doi:10.1093/cid/ciaa149
Valenzuela Nieto, G., Jara, R., Watterson, D., Modhiran, N., Amarilla, A. A., Himelreichs, J., et al. (2021). Potent Neutralization of Clinical Isolates of SARS-CoV-2 D614 and G614 Variants by a Monomeric, Sub-nanomolar Affinity Nanobody. Sci. Rep. 11 (1), 3318. doi:10.1038/s41598-021-82833-w
Valle, C., Martin, B., Touret, F., Shannon, A., Canard, B., Guillemot, J. C., et al. (2020). Drugs against SARS‐CoV ‐2: What Do We Know about Their Mode of Action? Rev. Med. Virol. 30 (6), 1–10. doi:10.1002/rmv.2143
Veggiani, G., and de Marco, A. (2011). Improved Quantitative and Qualitative Production of Single-Domain Intrabodies Mediated by the Co-expression of Erv1p Sulfhydryl Oxidase. Protein Expr. Purif. 79 (1), 111–114. doi:10.1016/j.pep.2011.03.005
Vu, K. B., Ghahroudi, M. A., Wyns, L., and Muyldermans, S. (1997). Comparison of Llama VH Sequences from Conventional and Heavy Chain Antibodies. Mol. Immunol. 34 (16-17), 1121–1131. doi:10.1016/s0161-5890(97)00146-6
Wagner, T. R., Ostertag, E., Kaiser, P. D., Gramlich, M., Ruetalo, N., Junker, D., et al. (2021). NeutrobodyPlex-monitoring SARS‐CoV‐2 Neutralizing Immune Responses Using Nanobodies. EMBO Rep. 22 (5), e52325. doi:10.15252/embr.202052325
Walter, J. D., Hutter, C. A. J., Zimmermann, I., Wyss, M., Egloff, P., Sorgenfrei, M., et al. (2020). Sybodies Targeting the SARS-CoV-2 Receptor-Binding Domain. BioRxiv. doi:10.1101/2020.04.16.045419
Wang, M.-Y., Zhao, R., Gao, L.-J., Gao, X.-F., Wang, D.-P., and Cao, J.-M. (2020). SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 10, 587269. doi:10.3389/fcimb.2020.587269
Wang, W., Xu, Y., Gao, R., Lu, R., Han, K., Wu, G., et al. (2020). Detection of SARS-CoV-2 in Different Types of Clinical Specimens. Jama 323 (18), 1843–1844. doi:10.1001/jama.2020.3786
Watanabe, Y., Allen, J. D., Wrapp, D., McLellan, J. S., and Crispin, M. (2020). Site-specific Glycan Analysis of the SARS-CoV-2 Spike. Science 369 (6501), 330–333. doi:10.1126/science.abb9983
Weinstein, J. B., Bates, T. A., Leier, H. C., McBride, S. K., Barklis, E., and Tafesse, F. G. (2022). A Potent Alpaca-Derived Nanobody that Neutralizes SARS-CoV-2 Variants. iScience 25 (3), 103960. doi:10.1016/j.isci.2022.103960
Weninger, A., Hatzl, A.-M., Schmid, C., Vogl, T., and Glieder, A. (2016). Combinatorial Optimization of CRISPR/Cas9 Expression Enables Precision Genome Engineering in the Methylotrophic Yeast Pichia pastoris. J. Biotechnol. 235, 139–149. doi:10.1016/j.jbiotec.2016.03.027
Wolter, N., Jassat, W., von Gottberg, A., Cohen, C., and Cohen, C. (2022). Clinical Severity of Omicron Sub-lineage BA.2 Compared to BA.1 in South Africa. medRxiv. doi:10.1101/2022.02.17.22271030
Woźniak, M., Płoska, A., Siekierzycka, A., Dobrucki, L. W., Kalinowski, L., and Dobrucki, I. T. (2022). Molecular Imaging and Nanotechnology-Emerging Tools in Diagnostics and Therapy. Ijms 23 (5), 2658. doi:10.3390/ijms23052658
Wrapp, D., De Vlieger, D., Corbett, K. S., Torres, G. M., Wang, N., Van Breedam, W., et al. (2020). Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 181 (5), 1004–1015. e15. doi:10.1016/j.cell.2020.04.031
Wrenn, J. O., Pakala, S. B., Vestal, G., Shilts, M. H., Brown, H. M., Bowen, S. M., et al. (2022). COVID‐19 Severity from Omicron and Delta SARS‐CoV‐2 Variants. Influenza Resp. Viruses. doi:10.1111/irv.12982
Wu, X., Cheng, L., Fu, M., Huang, B., Zhu, L., Xu, S., et al. (2021). A Potent Bispecific Nanobody Protects hACE2 Mice against SARS-CoV-2 Infection via Intranasal Administration. Cell Rep. 37 (3), 109869. doi:10.1016/j.celrep.2021.109869
Wu, Y., Jiang, S., and Ying, T. (2017). Single-Domain Antibodies as Therapeutics against Human Viral Diseases. Front. Immunol. 8, 1802. doi:10.3389/fimmu.2017.01802
Wu, Y., Li, C., Xia, S., Tian, X., Kong, Y., Wang, Z., et al. (2020). Identification of Human Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe 27 (6), 891–898. e5. doi:10.1016/j.chom.2020.04.023
Wu, Z., Hu, Y., Xu, M., Chen, Z., Yang, W., Jiang, Z., et al. (2021). Safety, Tolerability, and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine (CoronaVac) in Healthy Adults Aged 60 Years and Older: a Randomised, Double-Blind, Placebo-Controlled, Phase 1/2 Clinical Trial. Lancet Infect. Dis. 21 (6), 803–812. doi:10.1016/S1473-3099(20)30987-7
Xiang, Y., Nambulli, S., Xiao, Z., Liu, H., Sang, Z., Duprex, W. P., et al. (2020). Versatile and Multivalent Nanobodies Efficiently Neutralize SARS-CoV-2. Science 370 (6523), 1479–1484. doi:10.1126/science.abe4747
Xiaojie, S., Yu, L., Lei, Y., Guang, Y., and Min, Q. (2020). Neutralizing Antibodies Targeting SARS-CoV-2 Spike Protein. Stem Cell Res. 50, 102125. doi:10.1016/j.scr.2020.102125
Xie, Y. J., Dougan, M., Jailkhani, N., Ingram, J., Fang, T., Kummer, L., et al. (2019). Nanobody-based CAR T Cells that Target the Tumor Microenvironment Inhibit the Growth of Solid Tumors in Immunocompetent Mice. Proc. Natl. Acad. Sci. U.S.A. 116 (16), 7624–7631. doi:10.1073/pnas.1817147116
Xu, J., Xu, K., Jung, S., Conte, A., Lieberman, J., Muecksch, F., et al. (2021). Nanobodies from Camelid Mice and Llamas Neutralize SARS-CoV-2 Variants. Nature 595 (7866), 278–282. doi:10.1038/s41586-021-03676-z
Yao, H., Cai, H., Li, T., Zhou, B., Qin, W., Lavillette, D., et al. (2021). A High-Affinity RBD-Targeting Nanobody Improves Fusion Partner's Potency against SARS-CoV-2. PLoS Pathog. 17 (3), e1009328. doi:10.1371/journal.ppat.1009328
Ye, G., Gallant, J., Zheng, J., Massey, C., Shi, K., Tai, W., et al. (2021). The Development of Nanosota-1 as Anti-SARS-CoV-2 Nanobody Drug Candidates. eLife 10. doi:10.7554/eLife.64815
Yetisen, A. K., Akram, M. S., and Lowe, C. R. (2013). Paper-based Microfluidic Point-Of-Care Diagnostic Devices. Lab. Chip 13 (12), 2210–2251. doi:10.1039/c3lc50169h
Yoon, S., Kim, S., and Kim, J. (2010). Secretory Production of Recombinant Proteins in Escherichia coli. Biot 4 (1), 23–29. doi:10.2174/187220810790069550
Yuan, T. Z., Garg, P., Wang, L., Willis, J. R., Kwan, E., Hernandez, A. G. L., et al. (2022). Rapid Discovery of Diverse Neutralizing SARS-CoV-2 Antibodies from Large-Scale Synthetic Phage Libraries. MAbs 14 (1), 2002236. doi:10.1080/19420862.2021.2002236
Zhu, M., Hu, Y., Li, G., Ou, W., Mao, P., Xin, S., et al. (2014). Combining Magnetic Nanoparticle with Biotinylated Nanobodies for Rapid and Sensitive Detection of Influenza H3N2. Nanoscale Res. Lett. 9 (1), 528. doi:10.1186/1556-276X-9-528
Keywords: nanobodies (VHH), SARS-CoV-2, COVID-19, virus, llama (Lama glama), alpaca (Vicugna pacos), VHH antibody fragment
Citation: Valenzuela-Nieto G, Miranda-Chacon Z, Salinas-Rebolledo C, Jara R, Cuevas A, Berking A and Rojas-Fernandez A (2022) Nanobodies: COVID-19 and Future Perspectives. Front. Drug. Discov. 2:927164. doi: 10.3389/fddsv.2022.927164
Received: 23 April 2022; Accepted: 17 June 2022;
Published: 18 July 2022.
Edited by:
Juan Fernandez-Recio, Institute of Vine and Wine Sciences (CSIC), SpainReviewed by:
Fabian Glaser, Technion Israel Institute of Technology, IsraelCopyright © 2022 Valenzuela-Nieto, Miranda-Chacon, Salinas-Rebolledo, Jara, Cuevas, Berking and Rojas-Fernandez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alejandro Rojas-Fernandez, alejandro.rojas@uach.cl
 Guillermo Valenzuela-Nieto
Guillermo Valenzuela-Nieto Zaray Miranda-Chacon
Zaray Miranda-Chacon Constanza Salinas-Rebolledo
Constanza Salinas-Rebolledo Ronald Jara
Ronald Jara Alexei Cuevas
Alexei Cuevas Anne Berking
Anne Berking Alejandro Rojas-Fernandez
Alejandro Rojas-Fernandez