Immuno Informatic Analysis of B-Cell Epitope Changes in SARS-Cov-2 Variants with Dominant S-Protein Mutations
Article Information
Xianlin Yuan1, Liangping Li2*
1Department of Food and Biological Engineering, Guangdong Industry Technical College, Guangzhou 510300, China
2Department of Oncology and Institute of Clinical Oncology, The first Affiliated Hospital, Jinan University, Guangzhou, Guangdong, People’s Republic of China.
*Corresponding author: Liangping Li, Department of Oncology and Institute of Clinical Oncology, The first Affiliated Hospital, Jinan University, Guangzhou, Guangdong, People’s Republic of China.
Received: 07 November 2021; Accepted: 19 November 2021; Published: 13 December 2021
Citation: Li L, Yuan X. Immuno informatic analysis of B-cell epitope changes in SARS-CoV-2 variants with dominant S-protein mutations. Journal of Bioinformatics and Systems Biology 4 (2021): 162-181.
View / Download Pdf Share at FacebookAbstract
Background Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV)-2 has been transmitted worldwide and resulted in the coronavirus disease 2019 (COVID-19) pandemic more than one year. The Spike protein (S protein) on the virus surface has shown several variants, which may influence viral antigenicity and vaccine efficacy. Methods: we used bioinformatics tools to analyze the B-cell epitopes of the prototype S protein and its nine common variants.
Results Twelve potentially linear and 53 discontinuous epitopes of B-cells were predicted from the prototype S protein. By comparing the epitope alterations between the prototype S protein and its variants, we found that the B-cell epitopes of these 11 variants had significantly different alterations. The D614G variant impacted the potential epitope only with moderately increased antigenicity, whereas the epitopes and antigenicity of some new dominant variants (e.g., E484k, N501Y) changed greatly.
Conclusion These results suggest that currently developed vaccines should be valid for SARS-CoV-2 infections with few epitope alterations, but there is a risk of reducing vaccine reactivity for variants with multiple altered epitopes and antigenicity. This study provides a rapid forecasting method for SARS-CoV-2 S protein eptitope changes and for taking precautions against the probable appearance of antigen escape induced by genetic variations of SARS-CoV-2.
Keywords
Mutation; Neutralizing antibody; Receptor-binding domain; SARS-CoV-2; Spike protein; Vaccine
Mutation articles; Neutralizing antibody articles; Receptor-binding domain articles; SARS-CoV-2 articles; Spike protein articles; Vaccine articles
Mutation articles Mutation Research articles Mutation review articles Mutation PubMed articles Mutation PubMed Central articles Mutation 2023 articles Mutation 2024 articles Mutation Scopus articles Mutation impact factor journals Mutation Scopus journals Mutation PubMed journals Mutation medical journals Mutation free journals Mutation best journals Mutation top journals Mutation free medical journals Mutation famous journals Mutation Google Scholar indexed journals Neutralizing antibody articles Neutralizing antibody Research articles Neutralizing antibody review articles Neutralizing antibody PubMed articles Neutralizing antibody PubMed Central articles Neutralizing antibody 2023 articles Neutralizing antibody 2024 articles Neutralizing antibody Scopus articles Neutralizing antibody impact factor journals Neutralizing antibody Scopus journals Neutralizing antibody PubMed journals Neutralizing antibody medical journals Neutralizing antibody free journals Neutralizing antibody best journals Neutralizing antibody top journals Neutralizing antibody free medical journals Neutralizing antibody famous journals Neutralizing antibody Google Scholar indexed journals Receptor-binding domain articles Receptor-binding domain Research articles Receptor-binding domain review articles Receptor-binding domain PubMed articles Receptor-binding domain PubMed Central articles Receptor-binding domain 2023 articles Receptor-binding domain 2024 articles Receptor-binding domain Scopus articles Receptor-binding domain impact factor journals Receptor-binding domain Scopus journals Receptor-binding domain PubMed journals Receptor-binding domain medical journals Receptor-binding domain free journals Receptor-binding domain best journals Receptor-binding domain top journals Receptor-binding domain free medical journals Receptor-binding domain famous journals Receptor-binding domain Google Scholar indexed journals SARS-CoV-2 articles SARS-CoV-2 Research articles SARS-CoV-2 review articles SARS-CoV-2 PubMed articles SARS-CoV-2 PubMed Central articles SARS-CoV-2 2023 articles SARS-CoV-2 2024 articles SARS-CoV-2 Scopus articles SARS-CoV-2 impact factor journals SARS-CoV-2 Scopus journals SARS-CoV-2 PubMed journals SARS-CoV-2 medical journals SARS-CoV-2 free journals SARS-CoV-2 best journals SARS-CoV-2 top journals SARS-CoV-2 free medical journals SARS-CoV-2 famous journals SARS-CoV-2 Google Scholar indexed journals Spike protein articles Spike protein Research articles Spike protein review articles Spike protein PubMed articles Spike protein PubMed Central articles Spike protein 2023 articles Spike protein 2024 articles Spike protein Scopus articles Spike protein impact factor journals Spike protein Scopus journals Spike protein PubMed journals Spike protein medical journals Spike protein free journals Spike protein best journals Spike protein top journals Spike protein free medical journals Spike protein famous journals Spike protein Google Scholar indexed journals Vaccine articles Vaccine Research articles Vaccine review articles Vaccine PubMed articles Vaccine PubMed Central articles Vaccine 2023 articles Vaccine 2024 articles Vaccine Scopus articles Vaccine impact factor journals Vaccine Scopus journals Vaccine PubMed journals Vaccine medical journals Vaccine free journals Vaccine best journals Vaccine top journals Vaccine free medical journals Vaccine famous journals Vaccine Google Scholar indexed journals genome sequence articles genome sequence Research articles genome sequence review articles genome sequence PubMed articles genome sequence PubMed Central articles genome sequence 2023 articles genome sequence 2024 articles genome sequence Scopus articles genome sequence impact factor journals genome sequence Scopus journals genome sequence PubMed journals genome sequence medical journals genome sequence free journals genome sequence best journals genome sequence top journals genome sequence free medical journals genome sequence famous journals genome sequence Google Scholar indexed journals pathogen articles pathogen Research articles pathogen review articles pathogen PubMed articles pathogen PubMed Central articles pathogen 2023 articles pathogen 2024 articles pathogen Scopus articles pathogen impact factor journals pathogen Scopus journals pathogen PubMed journals pathogen medical journals pathogen free journals pathogen best journals pathogen top journals pathogen free medical journals pathogen famous journals pathogen Google Scholar indexed journals Phylogenetic analysis articles Phylogenetic analysis Research articles Phylogenetic analysis review articles Phylogenetic analysis PubMed articles Phylogenetic analysis PubMed Central articles Phylogenetic analysis 2023 articles Phylogenetic analysis 2024 articles Phylogenetic analysis Scopus articles Phylogenetic analysis impact factor journals Phylogenetic analysis Scopus journals Phylogenetic analysis PubMed journals Phylogenetic analysis medical journals Phylogenetic analysis free journals Phylogenetic analysis best journals Phylogenetic analysis top journals Phylogenetic analysis free medical journals Phylogenetic analysis famous journals Phylogenetic analysis Google Scholar indexed journals
Article Details
1. Introduction
A severe, contagious type of pneumonia caused by a novel coronavirus was first reported in Wuhan (Hubei Province, China) in December 2019 and was detected in other countries shortly afterwards [1]. This disease was named “coronavirus disease 2019” (COVID-19) by the World Health Organization (WHO) on 11 April 2020. The genome sequence of the pathogen was soon identified with next-generation sequencing by the team of Professor Zhang Yongzhen at Shanghai Public Health Clinical Center &School of Public Health, Fudan University, Shanghai, China (accession number: MN908947 from the National Center for Biotechnology Information (NCBI) database) to be a novel beta-coronavirus belonging to the family Coronaviridae. The genome of this coronavirus and pathology of COVID-19 was similar to that of the severe acute respiratory syndrome coronavirus (SARS-CoV) infection that broke out in 2002 [2], so this coronavirus was named “SARS-CoV-2”.
Phylogenetic analysis [1] revealed SARS-CoV-2 to share 79.6% sequence identity with SARS-CoV [3] and 50% with the Middle-East respiratory syndrome coronavirus (MERS-CoV) 4]. Though it has a similar genome, SARS-CoV-2 is far more contagious, spreads much faster, and is more destructive than SARS-CoV [5] and MERS-CoV [6]. On 30 January 2020, the WHO announced the COVID-19 contagion to be a worldwide public-health emergency. As of 5 November 2021, 248,467,363 confirmed cases of COVID-19 including 5,027,183 deaths have been reported globally according to COVID-19 Weekly Epidemiological Update report of WHO(website at https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports). The virion of SARS-CoV-2 is spherical, enveloped, 60–140 nm in diameter, with spikes of about 9–12 nm protruding from its surface. The SARS-CoV-2 genome encodes 10 proteins, four of which are major structural proteins: Spike (S), Membrane (M), Envelope (E) and nucleocapsid (N) [7]. Each of these proteins is responsible for different functions in the SARS-CoV-2 life cycle. The M protein decides the shape and pattern of the virus envelope. Viral assembly and germination is accomplished by the E protein. The N protein and the RNA genome of the virion are closely linked and participate in the replication and assembly of SARS-CoV-2. Most importantly, the S protein acts as a “bridge” to attach and bind to the cell receptors of the host. This action results in the fusion of the cellular membranes of the virus and host, and SARS-CoV-2 entry into the host cell [7]. The S protein is a type-I Transmembrane (TM) glycoprotein. It is composed of an ectodomain, TM domain and Cytoplasmic (CT) domain. The ectodomain is made up of two subunits (S1 and S2). The S1 subunit includes the N-terminal domain (NTD) and receptor-binding domain (RBD). The S2 subunit contains a fusion peptide and heptad repeat domains 1 and 2. The RBD is responsible for binding to the receptors of angiotensin-converting enzyme 2 in host cells. The S2 subunit completes viral fusion and entry into host cells [8]. Studies on SARS-CoV have shown that the RBD is a major target of effective neutralizing antibodies. Therefore, the S protein is not only a trigger of the replication and transmission of the virus, it is also the key target of a SARS-CoV-2 vaccine for COVID-2019 prevention.
However, owing to the extensive transmission of SARS-CoV-2, its genetic variants have appeared in an increasing number of countries. As of 1 May 2020, 5775 mutations in the SARS-CoV-2 genome were documented from 10,022 public genome-data assemblies [9], in which 394 missense mutations of the S protein were detected. Among these spike mutations, the D614G mutation (in which aspartic acid was replaced with glycine at the amino acid (AA) residue of 614) was a major mutation of great concern [10,11]. SARS-CoV-2 with the D614G mutation may have triggered fatal infections in Spain, Italy, and France [11]. It has been reported that the new virus variant N501Y had already reached 1/4 of the total cases of infection and 66.7% of cases infected in Britain in December 2020. At this rate, the spreading rate of N501Y variant maybe greatly more than the SARS-CoV-2 prototype [12]. These mutations will undoubtedly cause changes in the structure of the S protein. However, whether these mutations affect the antigenicity of the S protein and the binding ability with neutralizing antibodies is not known. If the B-cell epitopes on the S protein change and cannot not bind neutralizing antibodies, it would result in a loss of efficacy of the developed vaccine based on the prototype S protein. Several immuno-bioinformatics tools have been developed to analyze viral antigens, including linear and discontinuous epitopes of B-cells, as well as their immunogenicity. To explore these questions, we report use of such immuno-bioinformatics tools from the Immune Epitope Database (IEDB) and related resources to predict the B-cell epitopes of the S protein from the prototype and mutated strains of SARS-CoV-2. We also compared the changes of the likely epitope sites from dominant and rare mutations of the S protein. We found that the distinctive mutations of the S protein could affect the potentially effective epitopes of the S protein to different degrees.
1. Materials and Methods
1.1. Data retrieval and analyses of the number of variant
The primary sequence of the S protein of SARS-CoV-2 was retrieved from the NCBI GenBank database using accession number QHO62107.1. It has been used as the prototype sequence or reference sequence for vaccine development in many projects [13]. The accession number of its complete genome is NC_045512. The major variation sequences are available from the Global Initiative for Sharing All Influenza Data (GISAID) [14] and GenBank database. Just as for epidemiologic statistics on the number of mutations, we selected data collected from two time points to observe the change in the number of mutations. One dataset collected until 13 April 2020 was reported by Korber and colleagues in a pre-print publication [15], whereas another until 1 May 2020 was from Koyama and colleagues [9]. We undertook a secondary classification analysis on the 10,022 sequences listed in the supplementary data of the study by Koyama and colleagues, and obtained the number of different mutations of the S protein to calculate the mutation frequency. Then, we selected 10 variants with >10 mutations for further analyses.
1.2. Conservation analyses of selected sequences of the S protein
The sequences of the S protein from 10 countries were obtained randomly from an open NCBI Genbank database. By utilizing Clustal Omega tool v1.2.4 [16] and MSAViewer tool from Virus Pathogen Database and Analysis Resource (www. viprbrc.org), multiple sequence alignment (MSA) was carried out to perceive the conservation of sequences twice [17]. The MSAViewer tool could provide the visual-comparison results. The aligned files were also applied to create a phylogenetic tree via Clustal Omega Self-contained analytical tools (www.ebi.ac.uk/Tools).
1.3. Analyses of structure and antigenicity
First, we analyzed the secondary structure of the S protein of SARS-CoV-2. The Conserved Domain Database tool [18] in the NCBI website was used to analyze the main functional domains of the S protein, and to determine the detailed functional domains of the S protein with reference to the study of Lan and colleagues [19]. The TMHMM online tool (www.cbs.dtu.dk/ services/TMHMM/) was used to examine the TM topology of the S protein [20]. Homologous modeling of the S protein was carried out using the Swiss-model tool (https://swissmodel.expasy.org) to find its three-dimensional (3D) structural data. The confirmed 3D structure of the S protein of SARS-CoV-2 via electron microscopy at 3.2 Å was acquired using the Protein Data Bank (PDB ID: 6VYB) [8]. The antigenicity of the S protein was predicted by Vaxijen 2.0 at a default threshold of 0.4. This tool was developed to define antigen classification in view of the physicochemical properties of proteins rather than sequence alignment. It is now is a common tool for antigenicity assessment for vaccine design (http://www.ddg-pharmfac.net/vaxijen/) [21,22].
1.4. Prediction of the B-cell epitope of the S protein
We used the sequence from SARS-CoV-2 reported at the start of the COVID-19 epidemic as the wild-type or prototype, and the recent variants of SARS-CoV-2 as mutation strains, to predict the B-cell epitopes of the S protein. The S-protein sequence was exclusive of the Signal Peptide (SP), TM and CT regions, and only the ectodomain of the S protein was used for analyses. The linear and non-linear (discontinuous) epitopes of B cells were predicted using different tools. Linear epitopes were predicted by the BepiPred-2.0 server of IEDB (http://www.iedb.org/) [23,24]. The threshold was set to 0.55, which denoted a sensitivity of 29%, and specificity of 81%. Analytical results showed a figure in which the residues with scores above the threshold predicted to be part of an epitope were colored yellow. Effective B-cell epitopes rely on a stronger antigenicity and accessibility on the surface [25]. Then, total antigenicity scores were evaluated by Vaxijen 2.0 and the epitope surface accessibility was assessed through the Emini Surface Accessibility Prediction tool [26].The prediction of discontinuous epitopes was dependent upon surface accessibility, AA statistics, and the X-ray crystallography of protein epitopes [27]. We predicted the discontinuous epitopes of the prototype S protein via the DiscoTope 2.0 server [28] by entering the PDB ID number: 6VYB. The threshold was determined at 0.5, which manifested 23% sensitivity and 90% specificity.
1.5. Comparison of B-cell epitopes between the prototype S protein and mutated S protein
According to the research of Korber and colleagues [15] and Koyama and coworkers [9], we selected nine variants of the mutated S protein for analyses of their B-cell epitopes. Next, we compared each of them with the epitopes from the sequence of the prototype S protein to determine the influence of the mutation on epitopes. Finally, we summarized and listed all of major changes in a table.
1.6. Availability of data and codes
All data retrieved and analyzed in the present study were obtained from the NCBI, IEDB, GISAID and PDB and other open databases. The literature includes all quoted or analyzed data during this study, and are summarized in the figures, tables and Supplemental Information.
1.7. Statistical analyses
Statistical analyses were not applied in this theoretical study. Results are based on data in the literature and publicly available databases.
2. Results
2.1. Epidemiology statistics of S-protein mutations
First, we searched and compared the epidemiology statistical data of S-protein mutations published by Korber and colleagues as well as Koyama and coworkers. Korber and colleagues collected 4,535 genome sequences as of 13 April 2020; Koyama identified 5775 distinct genome variants from 10,022 SARS-CoV-2 genomes submitted to databases before 1 May 2020 [15]. In the report of Koyama and colleagues, the S protein contained 394 missense mutations [9]. In both reports, the mutation D614G had the highest frequency and other mutations were relatively rare. Quantitative statistical analyses of the major variants at two time points revealed that, in just 2 weeks, the number of D614G mutations had doubled, which demonstrated that this variant was the dominant form among S-protein mutations. The frequency of other mutations changed slightly, and some showed zero growth (Table 1). Several new variants that emerged in the fall of 2020 are most notably concerning. The information of these variants was obtained from the website of The Centers for Disease Control and Prevention (CDC), USA.
|
S protein mutation |
Amount of samples @ |
Amount of |
Domain |
Reference No. |
|
|
4/13/2020 |
samples* 05/1/2020 |
||||
|
L5F |
37 |
59 |
SP |
EPI_ISL_418992 # |
|
|
H49Y |
12 |
25 |
NTD |
EPI_ISL_403937 # |
|
|
Y145H |
10 |
12 |
NTD |
EPI_ISL_417665 # |
|
|
V367F |
12 |
15 |
RBD |
EPI_ISL_411219 # |
|
|
G476S |
8 |
10 |
RBD |
EPI_ISL_417085 # |
|
|
V483A |
21 |
26 |
RBD |
EPI_ISL_417159 # |
|
|
E484k |
283^ |
RBD |
|||
|
N501Y |
97955^ |
RBD |
|||
|
D614G |
3577 |
6294 |
S1 and S2 |
EPI_ISL_406862 # |
|
|
connection |
|||||
|
V615I/F |
13 |
13 |
S1 and S2 |
MT326090 & |
|
|
connection |
|||||
|
A831V |
28 |
27 |
S2 |
EPI_ISL_424384 # |
|
|
P1263L |
49 |
50 |
CT |
EPI_ISL_433679 # |
|
|
@Statistics from Korber B et al.’s study[15] |
|||||
Table 1:Distribution and statistics of the common mutations on S protein.
2.2. Conservation analyses of the prototype S protein
We chose the earliest submitted strain (NCBI ID:QHO62107.1) detected in China as the prototype S protein. Before predicting B-cell epitopes, we carried out conservation analyses by comparing this sequence with other, earlier sequences submitted from different countries in January–February 2020. The S-protein sequence of SARS-CoV-2 from the isolates of 10 countries (China (NCBI ID:QHO62107.1), Japan (BBW89517.1), USA (QHQ82464.1), Germany (QKM76570.1), Egypt (QKS66892.1), Spain (QKJ68388.1), France (QJT72638.1), Greece (QIZ16535.1), Australia (QHR84449.1) and Russia (QKV28206.1)) were subjected to MSA through the Clustal Omega tool (Figure S1). Conservation analyses of S-protein sequences demonstrated that the prototype S protein had 100% identity with all retrieved sequences. Thus, we used the earliest version of the S-protein sequence as the prototype for subsequent prediction of B-cell epitopes.
2.3. Structural analyses of the prototype S protein
We wished to localize these mutations in the position of different functional domains of the S protein. We analyzed S-protein sequences using bioinformatics tools. To analyze the topology of TM proteins, we applied the online tool TMHMM to treat the sequence of the S protein, and localized one TM region. The spatial distribution of residues could be divided into three parts: residues from 1 to 1213 on the extracellular surface; residues from 1214 to 1236 in the TM region; residues from 1237 to 1273 in the CT region. The extracellular domains divide into S1 and S2 subunits; S1 contains the NTD and RBD [19]. Based on the beginning and end positions of different domains, 10 major mutations were shown in the schematic diagram of the S protein (Figure 1A). Most of the S-protein mutations (70%) were located in the S1 subunit and the junction between S1 and S2 subunit. The most frequent mutation, D614G, was near the RBD. We wished to show functional domains at the 3D level and to analyze the discontinuous epitopes of the S protein. We searched databases through homology modeling of the Swiss-model tool. We found the 3D-structure file of the S protein (PDB ID: 6vyb) in which the AA sequences were 99.5% consistent with the S protein. The S protein of SARS-CoV-2 is a trimer, and the 3D structure of the ectodomain (open state) from 6vyb is shown in Figure 1B [13]: it contains three chains of A/B/C.
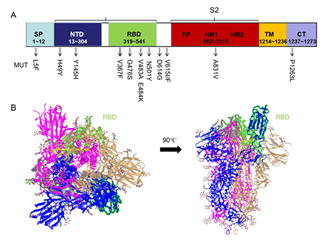
Figure 1: RBD (indicated in green highlight) is shown in iCn3D Viewer.
2.4. Prediction of B-cell epitopes on the prototype S protein
Humoral immunity has a very important role in defense against viral infection. The B-cell receptor or neutralizing antibodies recognize the B-cell epitopes (linear and discontinuous) of the S protein which, in general, exist on the SARS-CoV-2 surface as non-processed, natural antigen molecules. To predict the potential linear B-cell epitopes, we first used the BepiPred-2.0 prediction tool on IEDB to screen the prototype S-protein sequence: we discovered 30 B-cell linear epitopes (Table S3), the distribution of which is shown in (Figure 2A). Most of the B-cell epitopes were located on the NTD and RBD of the S protein. Next, we identified the effective epitopes by analyzing the antigenicity with the Vaxijen 2.0 tool and accessibility with the Emini Surface Accessibility Prediction tool (Figure 2B). Twelve effective epitopes were found from 30 predicted epitopes. The position, sequence, length and evaluation scores of potential B-cell linear epitopes are listed in Table 2. Among them, nine epitopes were in the S1 subunit (four in the NTD and five in the RBD) and three epitopes in the S2 subunit of the S protein. Based on this analysis, we found that three epitopes in the RBD (384PTKLNDL390, 405DEVRQIAPGQTGKI418, and 487NCYFPL492) had more significant antigenicity and accessibility compared with other epitopes.
We also predicted discontinuous epitopes using the Discotope 2.0 online server. The 3D structure of the S protein (PDB ID: 6vyb; Chain ID: A) was utilized to predict the discontinuous epitopes. The default threshold was −3.7 with 47% sensitivity and 75% specificity. The 53 discontinuous epitopes were predicted and located mainly in the whole RBD at 400 AAs to 600 AAs of the S protein (Figure 3A). All of the predicted epitopes distributed on the surface of the S protein are shown in 3D in Figure 3B using JSmol Viewer. According to their distribution in different domains, these epitopes (Table S4) could be divided into four groups (Table 3). The highest propensity score and DiscoTope score of epitopes were concentrated at 498 AAs to 500 AAs of the RBD (arrows in Figure 3B). Among them ,501 belongs to the conformational epitope, which indicates that the epitope of the spatial conformation has a great influence on the N501Y mutation of the new variant. Finally, these epitopes were validated by the Pepitope tool (http://pepitope.tau.ac.il/). The three major antigen clusters were consistent with the B-linear epitopes mentioned above (Table 4), which further indicated the rationality of our predicted B-linear epitopes.
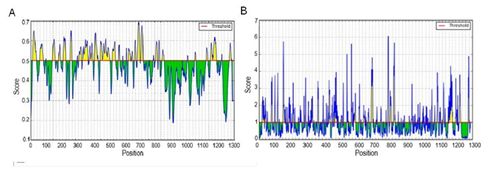
Figure 2: Prediction of B-cell linear epitopes and accessibility analyses of the prototype S protein.
(A) Distribution of all the predicted B-cell linear epitopes by BepiPred-2.0. The residues with scores above the threshold (value is adjusted at 0.55) are predicted to be potential epitopes and colored yellow. The Y-axis indicates residue scores and X-axis denotes the amino-acid-residue positions of the S protein. (B) Surface accessibility analyses using the Emini Surface Accessibility Scale. The residues with scores above the threshold (default value is 1.00) are predicted to have good accessibility.
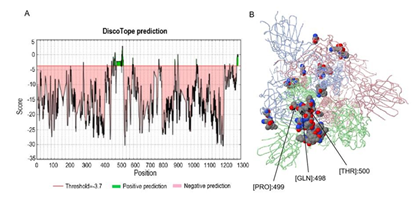
Figure 3: Prediction and distribution of B-cell discontinuous epitopes of the prototype S protein.
The discontinuous epitopes of the S protein were predicted by the Discotope 2.0 tool and the default threshold was -3.7. (A) The distribution of the predicted discontinuous epitopes in the prototype S protein. The green part in the figure represents the possible presence of discontinuous epitopes from the RBD, whereas the pink region indicates unlikely epitopes. (B) The surface position of discontinuous antigen epitopes in the 3D structure of the prototype S protein. The arrows in the figure refer to residues from the RBD with the highest DiscoTope score, which could have the potential to induce a better immune response.
|
No. |
Position |
Peptde |
Length 10 |
Accessibility |
Antigenicity |
Domain |
|
1 |
15~31 |
CVNLTTRTQLPPAYT NS |
17 |
4.781 |
1.2219 |
NTD |
|
2 |
62~75 |
VTWFHAIHVSGTNG |
14 |
1.114 |
0.5786 |
NTD |
|
3 |
141~152 |
LGVYYHKNNKSW |
12 |
4.321 |
0.8156 |
NTD |
|
4 |
208~220 |
TPINLVRDLPQGF |
13 |
1.534 |
0.4768 |
NTD |
|
5 |
384~390 |
PTKLNDL |
7 |
2.602 |
1.0882 |
RBD |
|
6 |
405~418 |
DEVRQIAPGQTGKI |
14 |
2.716 |
0.9312 |
RBD |
|
7 |
441~448 |
LDSKVGGN |
8 |
1.156 |
0.8773 |
RBD |
|
8 |
459~464 |
SNLKPF |
6 |
1.239 |
0.5943 |
RBD |
|
9 |
487~492 |
NCYFPL |
6 |
1.388 |
0.996 |
RBD |
|
10 |
657~664 |
NNSYECDI |
8 |
1.127 |
0.6539 |
S2 |
|
11 |
696~709 |
TMSLGAENSVAYSN |
14 |
1.995 |
0.678 |
S2 |
|
12 |
1154~1169 |
KYFKNHTSPDVDLGD I |
16 |
4.097 |
0.7333 |
S2 |
Table 2: The selected B-cell linear epitopes and evaluation scores.
|
No. |
Discontinuous epitopes |
Domain |
|
|
1 |
ASN282 |
NTD 13~304 |
|
|
2 |
THR415, ASN439, ASN449, ASN448, TYR449, ASN450, LEU455, PHE456,ARG457, LYS458, SER459, ASN460, LYS462, ILE468, SER469, THR470,PHE490, LEU492,GLN493, SER494, GLY496, GLN498, PRO499, THR500, VAL503,ASN 501, VAL503, GLY504,TYR505 |
RBD 319~541 |
|
|
3 |
ASN556, LYS558, LEU560, PRO561 |
S1 and S2 connection |
|
|
4 |
ASN703,SER704,VAL705, PRO793,ILE794,PRO809,SER810, LYS811, ASN914, TYR917, GLU918, PRO1140, LEU1141, GLN1142, PRO1143, GLU1144, LEU1145, ASP1146,SER1147 |
S2 662~1213 |
|
|
Residues in the epitopes that have the highest P-Score and D-Score are underlined. The site of the new variant N501Y was highlighted in red. |
|||
Table 3: B cell Discontinuous epitopes predicted via Discotope 2.0 tool.
|
Rank |
Residues number |
constitution |
Score |
|
Cluster 1 |
8 |
PHE377, SER383, PRO384, THR385, LYS386, ASN388, ASP389, LEU390 |
19.965 |
|
Cluster 2 |
14 |
THR696, MET697, SER698, LEU699, GLY700, ALA701, GLU702, ASN703, SER704, VAL705, ALA706, TYR707, SER708, ASN710 |
18.403 |
|
Cluster 3 |
12 |
THR208,PRO209,ILE210,ASN211,LEU212,VAL2 13,ARG214,ASP215,PRO217,GLN218,GLY219,PHE220 |
16.668 |
Table 4: Antigen cluster predicted via Pepitope tool.
2.5. Prediction of B-cell epitopes of the major variants of the S protein
Mutations of the S protein can influence its structure and change B-cell epitopes for the neutralizing antibody. Missense mutations will result in changes of AA residues and may affect B-cell epitopes. To assess the impact of the dominant and rare mutations of the S protein to linear B-cell epitopes, we selected the S-protein mutations with >10 counts from these 394 missense mutations to predict the B-cell epitopes. Among the several common mutations, we focused the analysis of B-cell epitopes on the ectodomains of the S protein, except the L5F mutation in the SP and P1263L mutation in the intracellular region (which do not appear on the SARS-CoV-2 surface). Due to a lack of 3D-structural data on mutated S proteins, we could not predict their discontinuous epitopes of B-cells. Through analyzing the sites of eleven missense mutations, we found that these conformational epitopes did not contain any sites of common mutations. Hence, mutation of the S protein had only a slight effect on the conformational epitope of B cells. Thus, we predicted the linear B-cell epitope of 11 variants (as described below) and determined the changes of epitopes by comparison with the epitopes from the prototype S protein.
2.5.1. H49Y mutation
The H49Y mutation occurred mainly in China, but seems to be reducing in overall frequency currently (15). Assessment of its antigenicity and surface availability suggested that four epitopes had changed (Table S5). In brief, after the H49Y mutation, the S protein had 14 effective epitopes, two of which had better antigenicity than that of the original epitopes (sites 405–417 and 697–709), two of which were newly generated (sites 519–533 and 618–629) and the remaining 10 epitopes were the same as those without a mutation.
2.5.2. Y145H mutation
The Y145H mutation was observed in eight countries, and it is a descendant of a highly infectious delta variant, which seems to be 10% to 15% more infectious than Delta [15]. Using the screening methods stated above, we found that five altered sites had distinct influences on the likely epitopes (Table S6). With emergence of the Y145H mutation, the S protein had 13 effective epitopes: two were newly produced from the originally unlikely epitopes (site 618–625), three had better antigenicity than the original epitopes (sites 140–153, 459–465 and 657–663) and the remaining nine epitopes were conserved.
2.5.3. V367F mutation
The V367F mutation was present in Europe and Hong Kong, but appeared to be declining in overall global distribution [15]. Only six alterations had obvious effects on likely epitopes (Table S7): two previously effective epitopes (sites 208–220 and 487– 492) in replacement for 210~221 and 487~489 would lose their function one epitope was deleted (site 459–464) and two alterations (141–152→140–154 and 208– 220→210–221) reduced the antigenicity of the original epitopes greatly. However, the antigenicity of two previous epitopes (385–392 and 404–416) was enhanced. Notably, there were nine potential epitopes after the V367F mutation in which the overall immunogenicity was reduced.
2.5.4. G476S
Six changes affected the effective epitope directly (Table S8). First, the previously likely epitopes at sites 62–75 and 459–464 were deleted. Second, the antigenicity of the epitope at site 216–221 was decreased below the threshold at which the epitope could be effective. Third, the antigenicity of a formerly ineffective epitope was increased so that it became an effective epitope at site 314–321. Fourth, two epitopes (sites 372–374 and 384–390) fused into a new epitope (site 368–390), but its antigenicity was lower than that of the previous epitope. Fifth, three alterations of epitopes improved their antigenicity (sites 406–417,440–450 and 657–663). Finally, the epitope at site 486–492 had slightly reduced antigenicity. Therefore, nine effective epitopes were predicted after the G476S mutation, in which the number of epitope changes was the most, and the overall antigenicity was reduced.
2.5.5. V483A mutation
Thirteen changes influenced the potential B-cell epitope of the V483A variant significantly (Table S9). These major changes could be grouped into four categories. The first category was deletion (sites 62–75 and 487–492). The antigenicity of one epitope (site 210–221) was too low to be a suitable epitope. The second change was additional epitopes. The antigenicity of epitopes at sites 181–186, 342–353, 363–377 and 617–628 was upregulated above the threshold needed to become new epitopes. The third change was reduced antigenicity. The antigenicity of the original epitope (site 405–418) was decreased at site 405–413. The final change was improved antigenicity. Four epitopes (sites 379–389, 442–447, 458–463, and 698–709) increased their antigenicity. Therefore, there were 13 effective epitopes in total, and the overall antigenicity after mutation was increased compared with that of the prototype S protein.
2.5.6. E484K mutation
In the South African variant E484K with the site on the ACE2 binding interface, which influences protein folding or is less favorable for binding [29]. However,this mutation results in impaired recognition of neutralizing antibodies and spike proteins, thereby enhancing the pathogenicity of the variant [29]. As for E484K mutation, 12 effective epitopes and 10 changes were predicted (Table S134). Compared with the prototype, these changes can be divided into four categories (Table S10): Firstly, one likely epitope was changed as site 141-154 and its antigenicity was reduced. Secondly, two previously non-antigenic sites extended and become new potential antigenic epitopes at site of 179-186 and 370-376. Thirdly, two original antigenic epitopes losed or decreased antigenicity to be non-antigen sites (458-481 and 487-492) due to mutation. Fourthly, the antigenicity of 5 epitopes were increased at sites in 209-220, 249-259, 410-416, 439-447 and 657-663. Overall, after E484K mutation, the antigenicity of the new variant S protein increased and the responsiveness to the vaccine was predicted well.
2.5.7 N501Y mutation
Spike mutation N501Y is also reported in the new UK variant under investigation, which is from a different clade, known as B.1.1.7 [30]. Structural analysis data showed that this mutation is more conducive to the combination of pike and ACE2, thus accelerating transmission [31].This variant maybe associated with an increasing risk of death compared with other variants as indicated by Centers for Disease Control (https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html). In this variant, 11 effective epitopes were predicted (Table S11) and 7 changes different from the prototype appears. These major changes could be grouped into four categories: firstly, the antigenicity of two epitopes (site 62-75, 140-154) was decreased. Secondly, a novel epitope was produced in the mutant at site of 176-186. Thirdly, the antigenicity of two epitopes (214-221, 657-664) was increased. Lastly, two originally neighboring epitopes at 459-464 and 487-492 were completely missing. These changes suggest that the overall antigenicity of the mutated virus is greatly reduced, and it is speculated that the role of the vaccine may be at risk of down-regulation.
2.5.8 D614G mutation
The SARS-CoV-2 strains with the G614 form are more transmissible. If it entered a new area, it could become the dominant infection form rapidly according to epidemiologic analyses [15]. Therefore, we paid considerable attention to analyze the immunologic characteristics of the D614G mutation. We found 29 B-cell linear epitopes and 12 effective epitopes were predicted. Surprisingly, only one epitope changed slightly when compared with the prototype S protein. The B-cell epitope at site 657–664 of the prototype S protein was shortened by one AA in the D614G mutation. This action changed the epitope at site 657–663. This led to a slight increase in accessibility and antigenicity (by 36.8% and 25.6%, respectively), suggesting that the G614 mutant was more likely to bind to neutralizing antibodies and that the binding efficacy was also increased compared with those in the D614 stain. The remaining epitopes were consistent with the non-mutated S protein (Table S12).Importantly, three epitopes in the RBD (384PTKLNDL390, 405DEVRQIAPGQTGKI418, and 487NCYFPL492) were identical between D614 and G614 forms.
2.5.9 V615I mutation
After the V615I mutation of the S protein, only one change affected the potential epitope (at site 657–663), with a slight increase in antigenicity (Table S13). The rest of 11 epitopes were as same as reference sequences. Finally, 12 potent epitopes were predicted (identical to the number predicted for the D614G variant).
2.5.10 V615F mutation
The background of the V615F mutation was consistent with that of the V615I mutation. Twelve alterations affected the B-cell epitope directly other than the V615I mutation (Table S14). Specifically, the antigenicity of the four epitopes decreased (sites 140– 154, 209–212, 441–444 and 487–497), in which the antigenicity of 209–212 and 441– 444 were too low to have an effect. The original likely epitope at site 384–390 was deleted. However, the antigenicity of four epitopes improved at sites 404–418, 458– 466, 656–664 and 697–709. There are three neoantigen epitopes at sites 180–186, 214–221 and 374–389 with strong antigenicity. Ultimately, 12 potent epitopes were predicted.
2.5.11 A831V mutation
Although the A831V mutation is emerging only in Iceland as a single lineage currently, it is found in a potential fusion peptide in the S2 subunit [15], which affects the pathogenicity of SARS-CoV-2 directly. After the A831V mutation of the S protein, five potential epitopes were altered (Table S15), but the remaining epitopes were consistent with the prototype S protein. Only the antigenicity of the epitope at site 141– 153 was reduced, whereas the other four epitopes greatly improved their antigenicity (sites 405–417, 459–465 618–625 and 657–663), of which site 618–625 became a new effective epitope because the antigenicity increased beyond the threshold. Therefore, the S protein with the A831V mutation had 13 potential epitopes with increasing antigenicity.
2.6. Comparison of the changes in the epitopes of variants
We wished to investigate the influence of the 11 common mutations of the S protein on B-cell epitopes. We compared the predicted epitopes of the prototype S protein and mutant S protein, analyzed the association of epitope changes among mutations, and determined the influence of mutations on B-cell epitopes. The detailed information of changes in each mutation is listed in Table S16.
Some mutations did not change or only slightly changed B-cell epitopes, whereas others strongly impacted the number and site of B-cell epitopes. All the major changes in B-cells epitopes are summarized in Table 5. The most important finding is that the most common mutation, D614G, changed the B-cell epitopes of the S protein only slightly; it increased the accessibility and antigenicity of the epitope at site 657–663 only moderately. There were 12 potential epitopes in the D614G mutation, nearly identical to those without a mutation. There were only 12 effective epitopes of D614G and V615I mutations, in which only one epitope at the same site (657–663) could affect the potential effective epitopes with a slight increase in antigenicity. However, the number and forms of changes in epitopes were abundant in V483A, V615F, V367F,G476S, E484K and N501Y mutations. Among them, the alterations of epitopes in the V483A mutation were the most significant: 13 changes were present and 13 epitopes were predicted, and the antigenicity was highly improved. Three types of V367F, G476S and N501Y mutation were discovered to have reduced antigenicity as a whole. This change may lead to a decrease in the responsiveness of these mutants to the vaccine, and there is a potential risk of vaccine tolerance. In addition, the sites of change of some epitopes were common to several mutants. The change in epitopes at site 459–464 occurred in the mutations Y145H, V367F, G476S, V483A,N501Y,V615F and A831V. The epitope at site 657–663 was altered in Y145H, G476S, E484K, N501Y, D614G, V615I, V615F and A831V mutations. The epitope at site 1154–1169 did not change among the 11 variants.
Table 5: A comparison analysis of 11 mutations.
3. Discussion
Most COVID-19 vaccines have been designed to target the S protein to induce neutralizing antibodies [32,33]. Most vaccines entering phase-3 clinical trials are based on the S protein when SARS-CoV-2 was first sequenced [34,35]. However, the massive replication and rapid global transmission of SARS-CoV-2 has enabled it to mutate and evolve. By searching the literature and databases related to SARS-CoV-2, we found 11 common mutations of the S protein (Table 1) and five of them were concentrated on or near the RBD. The most frequently occurring mutation, D614G, was located at the junction of the S1 subunit and S2 subunit, which is near the furin cleavage site. Walls and colleagues reported that deletion of this cleavage site could influence SARS-CoV-2 S protein-mediated entry into host cells [8]. Hence, Korber and colleagues [15] and Zhang and coworkers [36] proposed that the D614G mutation contributed to the spread of SARS-CoV-2, which makes the G614 strain the dominant mutant.
A mutation in the S protein can affect B-cell epitopes and lead to vaccine failure. Therefore, to explore the impact of mutations on antigenicity of the S protein, we applied immuno-informatics tools to predict potential B-cell epitopes of the prototype S protein and variant S proteins. The reliably of prediction tools and methods has been demonstrated previously. For example, for MERS-CoV, Qamar and colleagues predicted the linear and discontinuous epitopes using the same immuno-informatics tools we employed in the present study [37]. In other experiments, the hMS-1,which is a monoclonal antibody isolated from MERS-CoV infected rehabilitators was shown to recognize some neutralizing epitopes at residues 510, 511 and 553 in RBD domain, and these discontinuous epitopes were predicted correctly from the work of Qamar and coworkers [38].
Our predicted epitopes of the prototype S protein coincided with the AA sites of neutralizing antibodies validated in other experimental studies. Cao and colleagues were the first to show that seven monoclonal antibodies isolated from 60 patients convalescing from COVID-19 showed strong binding affinity to the RBD and potent neutralizing ability against SARS-CoV-2 [39]. Those seven monoclonal antibodies possessed high structural similarity with m396 (shown previously to be a neutralizing antibody of SARS-CoV), which can recognize epitopes (sites 408, 442, 443, 460, 475) on the RBD of the S protein of SARS-CoV [40]. The RBD of the S protein is more prone to neutralizing antibodies [41], so we checked epitopes in the RBD (site 318– 550). Validation of the studies mentioned above was consistent with our predicted B-cell epitopes: five potential linear epitopes at AA residues 384–390, 405–418, 441– 448, 459–464 and 487–492 (Table 2). Also, discontinuous epitopes, such as RBD 319~541, especially GLN498, PRO499 and THR500 (Table 3) could be targets of vaccine candidates.
Importantly, whether mutations on the S protein leads to epitope changes merits investigation. We used a group of tools of B-cell epitopes to predict the prototype and variant S proteins. We demonstrated that an S protein with the D614G mutation had the least change of the potential effective B-cell epitopes, in which only one linear epitope had a slight change in the length of the non-RBD region compared with that in the prototype S protein. The predicted B-cell epitopes on the RBD were identical between D614 and G614 forms. Hence, this result suggested that an effective vaccine based on the prototype S protein should protect against infection by SARS-CoV-2 with the prototype S protein and D614G variant, which cover >90% of cases estimated from data of the study by Koyama and colleagues [9]. That is, the vaccines being developed currently could protect a large proportion of the SARS-CoV-2-infected population, including the D614 prototype and dominant G614 variant. Our prediction results are consistent with those of Weissman and colleagues [42], in which G614 spike was less susceptible to neutralization, but instead moderately. Those data suggest that the D614G mutation does not change B-cell epitopes to escape immune recognition. Otherwise, as viruses mutate, viruses actually evolve, including their ability to neutralize antibodies [43].Through our prediction, we observe that the virus is missing some original epitopes and producing new epitopes in the process of continuous variation,such as G476S, V483A, E484K, N501Y, V615F. This may be the immune defense mechanism of the virus in escaping human body and vaccine or antibody therapy.
Our prediction results also suggested that some mutations changed B-cell epitopes significantly (e.g., G476S, V483A, E484K and N501Y), which perhaps would influence the effect of a SARS-CoV-2 vaccine based on the prototype S protein. Use of the SARS-CoV-2 vaccine may produce selection pressure for these variants. These variants with more changes in potential epitopes may have significantly different responses to vaccines. If responsiveness to vaccines is reduced, it will filter the SARS-CoV-2 viral strains and appear transmission of drug resistance variants.
4. Conclusions
We demonstrated that the amount of S protein B cell epitope alteration caused by gene mutation could be rapidly predicted by immunobioinformatic analysis tools. These epitope changes may influence the effect of SARS-CoV-2 vaccine. Some mutations such as D614G slightly change B-cell epitopes, the published experimental data showed that the vaccine based on the prototype S protein could protect human populations against the infections caused by this variant of SARS-CoV-2. However, we also found that some mutations, e.g. N501Y, change B-cell epitopes significantly; these variants may lead to immune escape and be less responsive to vaccine. The bioinformatic analysis of virus variant immunogenicity should be a valuable approach to assess potential immune escape of new variants of SARS-CoV-2 to available vaccine.
Author Contributors
Xianlin Yuan undertook the analyses and wrote the paper. Liangping Li proposed and supervised this project, and wrote and revised the manuscript. All authors approved the final version of the manuscript.
Funding
This project was supported by National Key Research & Development Projects (2016YFC1303404) of the Ministry of Science and Technology of the People's Republic of China, and the Guangzhou Science and Technology Project (201604020133) from Guangzhou Science and Technology Innovation Commission.
Acknowledgments
We are grateful to Zijun Shu for his management of references in this manuscript. We also thank Charlesworth (https://www.cwauthors.com.cn) for its linguistic assistance during the polishing of this manuscript.
Declaration of interests
All authors declare no competing interests.
Supplementary materials
The supplementary material related with this study can be found in the attachment and in the online version.
https://www.fortunejournals.com/supply/JBSB-4517_file.pdf
References
- Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. nature 579 (2020): 270-273.
- Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New England journal of medicine 348 (2003): 1967-1976.
- Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell host & microbe 27 (2020): 325-328.
- Rabaan AA, Al-Ahmed SH, Haque S, et al. SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. Infez Med 28 (2020) 174-184.
- Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. New England journal of medicine 348 (2003) 1953-1966.
- Zaki AM, Van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine 367 (2012): 1814-1820.
- Malik YA. Properties of Coronavirus and SARS-CoV-2. The Malaysian Journal of Pathology 42 (2020): 3-11.
- Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell181 (2020):281-292.e6.
- Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bulletin of the World Health Organization. 98 (2020): 495-504.
- Becerra-Flores M, Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. International Journal of Clinical Practice 74 (2020): 13525.
- Eaaswarkhanth M, Al Madhoun A, Al-Mulla F. Could the D614 G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? International Journal of Infectious Diseases 96 (2020): 459-460.
- Conti P, Caraffa A, Gallenga CE, et al. The British variant of the new coronavirus-19 (Sars-Cov-2) should not create a vaccine problem.J Biol Regul Homeost Agents 35 (2020 ).
- Chen L, Liu W, Zhang Q, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerging microbes & infections 9 (2020a): 313-319.
- Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance 22 (2017): 30494.
- Korber B, Fischer WM, Gnanakaran S, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus.Cell 182 (2020): 812-827.
- Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences.Protein Sci 27 (2018 ): 135-145
- Yachdav G, Wilzbach S, Rauscher B, et al. MSAViewer: interactive JavaScript visualization of multiple sequence alignments. Bioinformatics. 32 (2016): 3501-3503.
- Lu S, Wang J, Chitsaz F, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48 (2020): 265-268.
- Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581 (2020):215-20.
- El-Rami FE, Sikora AE. Bioinformatics Workflow for Gonococcal Proteomics 1997. Methods Mol Biol (2019): 185-205.
- Doytchinova IA, Flower DR. Identifying candidate subunit vaccines using an alignment-independent method based on principal amino acid properties. Vaccine 25 (2007): 856-866.
- Conte FdP, Tinoco BC, Santos Chaves T, et al. Identification and validation of specific B-cell epitopes of hantaviruses associated to hemorrhagic fever and renal syndrome. PLoS neglected tropical diseases 13 (2019): 0007915.
- Peters B, Sidney J, Bourne P, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol 3 (2005): e91.
- Jespersen MC, Peters B, Nielsen M, et al. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic acids research 45 (2017): W24-W29.
- Fieser TM, Tainer JA, Geysen HM, et al. Influence of protein flexibility and peptide conformation on reactivity of monoclonal anti-peptide antibodies with a protein alpha-helix. Proceedings of the National Academy of Sciences 84 (1987): 8568-8572.
- Yao B, Zheng D, Liang S, et al. Conformational B-cell epitope prediction on antigen protein structures: a review of current algorithms and comparison with common binding site prediction methods. PloS one 8 (2013): 62249.
- Sun P, Ju H, Liu Z, et al. Bioinformatics resources and tools for conformational B-cell epitope prediction. Computational and mathematical methods in medicine (2013): 943636.
- Kringelum JV, Lundegaard C, Lund O. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol 8 (2012): 1002829.
- Jangra S, Ye C, Rathnasinghe R, et al. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera.medRxiv (2021).
- Leung K, Shum MH, Leung GM, et al. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 26 (2021): 2002106.
- Villoutreix B, Calvez V, Marcelin AG, et al. In silico investigation of the new UK (B.1.1.7) and South African (501Y.V2) SARS-CoV-2 variants with a focus at the ACE2-Spike RBD interface.Int J Mol Sci 22 (2021): 1695.
- Sharpe HR, Gilbride C, Allen E, et al. The early landscape of COVID-19 vaccine development in the UK and rest of the world. Immunology 160 (2020): 223-232.
- Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 12 (2020): 254.
- Dhama K, Sharun K, Tiwari R, et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Human Vaccines & Immunotherapeutics 16 (2020): 1232-1238
- Chen W, Strych U, Hotez PJ, et al. The SARS-CoV-2 Vaccine Pipeline: an Overview. Current tropical medicine reports (2020b): 1-4.
- Zhang L, Jackson CB, Mou H, et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity (2020).
- Qamar MTU, Saleem S, Ashfaq UA, et al. Epitope-based peptide vaccine design and target site depiction against Middle East Respiratory Syndrome Coronavirus: An immune-informatics study. Journal of Translational Medicine. 17 (2019): 1-14.
- Du L, Yang Y, Zhou Y, et al. MERS-CoV spike protein: a key target for antivirals. Expert opinion on therapeutic targets 21 (2017): 131-43.
- Cao Y, Su B, Guo X, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 182 (2020): 73-84.
- Zhu Z, Chakraborti S, He Y, et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Pro. Natl Acad Sci USA 104 (2007): 12123-12128
- Wong SK, Li W, Moore MJ, et al. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. Journal of Biological Chemistry 279 (2004). 3197-3201.
- Weissman D, Alameh MG, de Silva T, et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe 29 (2021): 23-31.
- Naqvi AAT, Fatima K, Mohammad T, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim Biophys Acta Mol Basis Dis 1866 (2020): 165878.


 Impact Factor: * 4.2
Impact Factor: * 4.2 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 