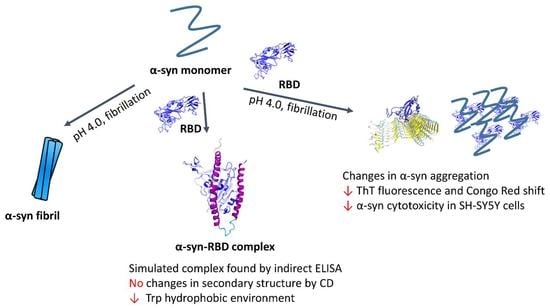
Does the SARS-CoV-2 Spike Receptor-Binding Domain Hamper the Amyloid Transformation of Alpha-Synuclein after All?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Purification of Recombinant Alpha-Synuclein
2.3. Preparation of Alpha-Synuclein Fibrils
2.4. Thioflavin T Fluorescence Assay
2.5. Fluorescence Spectroscopy
2.6. ANS Fluorescence
2.7. Congo Red Binding Assay
2.8. Modified ELISA for Studying Protein–Protein Interactions
2.9. Circular Dichroism
2.10. Cell Viability of Neuroblastoma SH-SY5Y Cells by MTT-Test
2.11. Docking and Molecular Dynamics Simulations
3. Results
3.1. Docking and Molecular Dynamics of Protein–Protein Interaction between SARS-CoV-2 Spike Receptor-Binding Domain (SARS-CoV-2 RBD) and Alpha-Synuclein (SYN)
3.2. Interaction of the Monomeric Form of Alpha-Synuclein with the SARS-CoV-2 RBD
3.3. Effect of SARS-CoV-2 RBD on the Amyloid Transformation of Alpha-Synuclein
3.4. Changes in Cytotoxicity of Synuclein Fibrils during Their Formation in the Presence of RBD
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mukaetova-Ladinska, E.B.; Kronenberg, G.; Raha-Chowdhury, R. COVID-19 and neurocognitive disorders. Curr. Opin. Psychiatry 2021, 34, 149–156. [Google Scholar] [CrossRef]
- De Luca, P.; Di Stadio, A.; Colacurcio, V.; Marra, P.; Scarpa, A.; Ricciardiello, F.; Cassandro, C.; Camaioni, A.; Cassandro, E. Long COVID, audiovestibular symptoms and persistent chemosensory dysfunction: A systematic review of the current evidence. Acta Otorhinolaryngol. Ital. 2022, 42, S87–S93. [Google Scholar] [CrossRef]
- De Felice, F.G.; Tovar-Moll, F.; Moll, J.; Munoz, D.P.; Ferreira, S.T. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and the Central Nervous System. Trends Neurosci. 2020, 43, 355–357. [Google Scholar] [CrossRef]
- Chatterjee, A.; Chakravarty, A. Neurological Complications Following COVID-19 Vaccination. Curr. Neurol. Neurosci. Rep. 2023, 23, 1–14. [Google Scholar] [CrossRef]
- Lingor, P.; Demleitner, A.F.; Wolff, A.W.; Feneberg, E. SARS-CoV-2 and neurodegenerative diseases: What we know and what we don’t. J. Neural. Transm. 2022, 129, 1155–1167. [Google Scholar] [CrossRef]
- Krey, L.; Huber, M.K.; Hoglinger, G.U.; Wegner, F. Can SARS-CoV-2 Infection Lead to Neurodegeneration and Parkinson’s Disease? Brain Sci. 2021, 11, 1654. [Google Scholar] [CrossRef]
- Silva, J.; Patricio, F.; Patricio-Martinez, A.; Santos-Lopez, G.; Cedillo, L.; Tizabi, Y.; Limon, I.D. Neuropathological Aspects of SARS-CoV-2 Infection: Significance for Both Alzheimer’s and Parkinson’s Disease. Front. Neurosci. 2022, 16, 867825. [Google Scholar] [CrossRef]
- Rahmani, B.; Ghashghayi, E.; Zendehdel, M.; Baghbanzadeh, A.; Khodadadi, M. Molecular mechanisms highlighting the potential role of COVID-19 in the development of neurodegenerative diseases. Physiol. Int. 2022, 109, 135–162. [Google Scholar] [CrossRef]
- Fu, Y.W.; Xu, H.S.; Liu, S.J. COVID-19 and neurodegenerative diseases. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 4535–4544. [Google Scholar]
- Sinha, S.; Mittal, S.; Roy, R. Parkinson’s Disease and the COVID-19 Pandemic: A Review Article on the Association between SARS-CoV-2 and alpha-Synucleinopathy. J. Mov. Disord. 2021, 14, 184–192. [Google Scholar] [CrossRef]
- Zenesini, C.; Vignatelli, L.; Belotti, L.M.B.; Baccari, F.; Calandra-Buonaura, G.; Cortelli, P.; Descovich, C.; Giannini, G.; Guaraldi, P.; Guarino, M.; et al. Risk of SARS-CoV-2 infection, hospitalization and death for COVID-19 in people with Parkinson’s disease or parkinsonism over a 15-month period: A cohort study. Eur. J. Neurol. 2022, 29, 3205–3217. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Rhea, E.M.; Logsdon, A.F.; Hansen, K.M.; Williams, L.M.; Reed, M.J.; Baumann, K.K.; Holden, S.J.; Raber, J.; Banks, W.A.; Erickson, M.A. The S1 protein of SARS-CoV-2 crosses the blood-brain barrier in mice. Nat. Neurosci. 2021, 24, 368–378. [Google Scholar] [CrossRef]
- Semerdzhiev, S.A.; Fakhree, M.A.A.; Segers-Nolten, I.; Blum, C.; Claessens, M. Interactions between SARS-CoV-2 N-Protein and alpha-Synuclein Accelerate Amyloid Formation. ACS Chem. Neurosci. 2022, 13, 143–150. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, X.; Huang, Z.; Ma, K. SARS-CoV-2 Proteins Interact with Alpha Synuclein and Induce Lewy Body-like Pathology In Vitro. Int. J. Mol. Sci. 2022, 23, 3394. [Google Scholar] [CrossRef]
- Idrees, D.; Kumar, V. SARS-CoV-2 spike protein interactions with amyloidogenic proteins: Potential clues to neurodegeneration. Biochem. Biophys. Res. Commun. 2021, 554, 94–98. [Google Scholar] [CrossRef]
- Salimian, J.; Ahmadi, A.; Amani, J.; Olad, G.; Halabian, R.; Saffaei, A.; Arabfard, M.; Nasiri, M.; Nazarian, S.; Abolghasemi, H.; et al. Safety and immunogenicity of a recombinant receptor-binding domain-based protein subunit vaccine (Noora vaccine) against COVID-19 in adults: A randomized, double-blind, placebo-controlled, Phase 1 trial. J. Med. Virol. 2022, 95. [Google Scholar] [CrossRef]
- Yang, J.; Liu, M.Q.; Liu, L.; Li, X.; Xu, M.; Lin, H.; Liu, S.; Hu, Y.; Li, B.; Liu, B.; et al. A triple-RBD-based mucosal vaccine provides broad protection against SARS-CoV-2 variants of concern. Cell. Mol. Immunol. 2022, 19, 1279–1289. [Google Scholar] [CrossRef]
- Greco, V.; Naletova, I.; Ahmed, I.M.M.; Vaccaro, S.; Messina, L.; La Mendola, D.; Bellia, F.; Sciuto, S.; Satriano, C.; Rizzarelli, E. Hyaluronan-carnosine conjugates inhibit Abeta aggregation and toxicity. Sci. Rep. 2020, 10, 15998. [Google Scholar] [CrossRef]
- Barinova, K.V.; Kuravsky, M.L.; Arutyunyan, A.M.; Serebryakova, M.V.; Schmalhausen, E.V.; Muronetz, V.I. Dimerization of Tyr136Cys alpha-synuclein prevents amyloid transformation of wild type alpha-synuclein. Int. J. Biol. Macromol. 2017, 96, 35–43. [Google Scholar] [CrossRef]
- Masuda, M.; Dohmae, N.; Nonaka, T.; Oikawa, T.; Hisanaga, S.; Goedert, M.; Hasegawa, M. Cysteine misincorporation in bacterially expressed human alpha-synuclein. FEBS Lett. 2006, 580, 1775–1779. [Google Scholar] [CrossRef] [Green Version]
- Niks, M.; Otto, M. Towards an optimized MTT assay. J. Immunol. Methods 1990, 130, 149–151. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.Y. HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jablonska, J.; Pravda, L.; Varekova, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. A Publ. Protein Soc. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- Yugandhar, K.; Gromiha, M.M. Protein-protein binding affinity prediction from amino acid sequence. Bioinformatics 2014, 30, 3583–3589. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Lee, S.; Tran, A.; Allsopp, M.; Lim, J.B.; Henin, J.; Klauda, J.B. CHARMM36 united atom chain model for lipids and surfactants. J. Phys. Chem. B 2014, 118, 547–556. [Google Scholar] [CrossRef]
- Yan, Y.; Huang, S.Y. Modeling Protein-Protein or Protein-DNA/RNA Complexes Using the HDOCK Webserver. Methods Mol. Biol. 2020, 2165, 217–229. [Google Scholar]
- Stroylov, V.S.; Svitanko, I.V.; Maksimenko, A.S.; Kislyi, V.P.; Semenova, M.N.; Semenov, V.V. Computational modeling and target synthesis of monomethoxy-substituted o-diphenylisoxazoles with unexpectedly high antimitotic microtubule destabilizing activity. Bioorganic Med. Chem. Lett. 2020, 30, 127608. [Google Scholar] [CrossRef]
- Agapova, Y.K.; Altukhov, D.A.; Timofeev, V.I.; Stroylov, V.S.; Mityanov, V.S.; Korzhenevskiy, D.A.; Vlaskina, A.V.; Smirnova, E.V.; Bocharov, E.V.; Rakitina, T.V. Structure-based inhibitors targeting the alpha-helical domain of the Spiroplasma melliferum histone-like HU protein. Sci. Rep. 2020, 10, 15128. [Google Scholar] [CrossRef] [PubMed]
- Medvedeva, M.; Barinova, K.; Melnikova, A.; Semenyuk, P.; Kolmogorov, V.; Gorelkin, P.; Erofeev, A.; Muronetz, V. Naturally occurring cinnamic acid derivatives prevent amyloid transformation of alpha-synuclein. Biochimie 2020, 170, 128–139. [Google Scholar] [CrossRef]
- Priss, A.; Afitska, K.; Galkin, M.; Yushchenko, D.A.; Shvadchak, V.V. Rationally Designed Protein-Based Inhibitor of alpha-Synuclein Fibrillization in Cells. J. Med. Chem. 2021, 64, 6827–6837. [Google Scholar] [CrossRef]
- Leisi, E.V.; Barinova, K.V.; Kudryavtseva, S.S.; Moiseenko, A.V.; Muronetz, V.I.; Kurochkina, L.P. Effect of bacteriophage-encoded chaperonins on amyloid transformation of alpha-synuclein. Biochem. Biophys. Res. Commun. 2022, 622, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Golts, N.; Snyder, H.; Frasier, M.; Theisler, C.; Choi, P.; Wolozin, B. Magnesium inhibits spontaneous and iron-induced aggregation of alpha-synuclein. J. Biol. Chem. 2002, 277, 16116–16123. [Google Scholar] [CrossRef]
- Nystrom, S.; Hammarstrom, P. Amyloidogenesis of SARS-CoV-2 Spike Protein. J. Am. Chem. Soc. 2022, 144, 8945–8950. [Google Scholar] [CrossRef]











Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stroylova, Y.; Konstantinova, A.; Stroylov, V.; Katrukha, I.; Rozov, F.; Muronetz, V. Does the SARS-CoV-2 Spike Receptor-Binding Domain Hamper the Amyloid Transformation of Alpha-Synuclein after All? Biomedicines 2023, 11, 498. https://doi.org/10.3390/biomedicines11020498
Stroylova Y, Konstantinova A, Stroylov V, Katrukha I, Rozov F, Muronetz V. Does the SARS-CoV-2 Spike Receptor-Binding Domain Hamper the Amyloid Transformation of Alpha-Synuclein after All? Biomedicines. 2023; 11(2):498. https://doi.org/10.3390/biomedicines11020498
Chicago/Turabian StyleStroylova, Yulia, Anastasiia Konstantinova, Victor Stroylov, Ivan Katrukha, Fedor Rozov, and Vladimir Muronetz. 2023. "Does the SARS-CoV-2 Spike Receptor-Binding Domain Hamper the Amyloid Transformation of Alpha-Synuclein after All?" Biomedicines 11, no. 2: 498. https://doi.org/10.3390/biomedicines11020498