Abstract
SARS-CoV-2 in vitro transcribed RNA reference materials (RM), UME RM 2019 and UME RM 2020, were produced by Scientific and Technological Research Council of Turkey (TUBITAK), National Metrology Institute (UME), to be used as a quality control material for SARS-CoV-2 measurements, in liquid-frozen and lyophilized forms, respectively. These RNA RMs include ten internationally recommended SARS-CoV-2 target gene fragments (Pasteur-RdRp-IP2, Pasteur-RdRp-IP4, Charite-E, Charite-RdRp, CDC-N1, CDC-N2, China CDC-ORF1ab, China CDC-N, Hong Kong-ORF1b, and Hong Kong-N) for virus detection and one human gene fragment (RNase P) as an internal control. Two different platforms, RT-qPCR and RT-ddPCR, were used to characterize UME RM 2019 (UME RM 2020 was only characterized with RT-qPCR). The homogeneity studies were evaluated by RT-qPCR. According to these results, it has been shown that both reference materials are homogeneous for intended use. Short-term studies were also conducted similarly for mimicking transport conditions and UME RM 2020, which is produced in lyophilized form, unlike other reference materials available in the market, provides convenience for users by ensuring that the reference material remains stable for 17 days even at 45 °C temperature. The lyophilized formulation of the reference material had greater stability which would allow it to be shipped without cooling items. The development of such RNA reference materials provides quality control for existing and newly designed RNA-based virus detection tests and it helps the prevention and control of epidemics.
Similar content being viewed by others
Introduction
COVID-19 (coronavirus disease-2019) is a respiratory disease that has killed more than one million people since its discovery in December 2019 [1]. This respiratory disease is caused by a new type of virus, SARS-CoV-2, from the Coronaviridae family. The SARS-CoV-2 virus is an enveloped, positive-sense, and single-stranded RNA virus. It is the seventh coronavirus reported to infect human species after SARS-CoV and MERS-CoV [2, 3]. This new type of coronavirus has been reported to be a member of the β coronavirus group like SARS-CoV and MERS-CoV [4]. SARS-CoV and MERS-CoV viruses from the Coronaviridae family have also caused respiratory tract infections and deaths in the past 20 years. The most important feature of the SARS-CoV-2, whose origin has not yet been clarified, is to spread rapidly from person-to-person through droplets and contact. Due to its high contagiousness, the spread of SARS-CoV-2 virus could not be controlled and the COVID-19 disease spread across the continents, turning it into a pandemic [5].
Early detection and isolation of cases infected with SARS-CoV-2 from the community are important to stop the spread of the disease and to control the pandemic. Reverse transcription real-time quantitative polymerase chain reaction (RT-qPCR) method is widely used for early detection of SARS-CoV-2 [6, 7]. This method consists of two steps: reverse transcription of ribonucleic acid (RNA) to complementary DNA (cDNA) and amplification of cDNA with PCR. Although RT-qPCR has been recognized as the gold standard by the World Health Organization (WHO) and the Chinese Center for Disease Control and Prevention (CCDC) [8, 9], it has certain limitations such as false positive and false negative results depending on the assay design. These limitations may result from sampling, PCR reagent factors (e.g., reverse transcriptase activity) [10], variable detection limit of test kits, and assay design. In order to improve the quality of measurements, it is important to use RNA reference materials in measurements used in the diagnosis of SARS-CoV-2 [11]. The use of RNA-based reference material is of great importance, especially when developing new assays and validating diagnostic kits. The reference materials can also be used to measure the sample viral load. To ensure the quality and reliability of the measurement results, reference materials are required [12].
As a result, it is possible to extend the limitations of RT-qPCR by using RNA-based reference materials. Using an RNA-based reference material will be able to contribute to the development of reliable and accurate diagnostic kits by controlling cDNA synthesis and PCR steps. Moreover, it will also be possible to determine and compare the detection limits of existing diagnostic kits. Only a few RNA-based reference materials are reported to be produced in liquid frozen [13,14,15]. Lyophilized form that enables shipping without any cooling item has not been reported. Therefore, The National Metrology Institute of Turkey (UME) functioning under Scientific and Technological Research Council of Turkey (TUBITAK) has produced lyophilized (UME RM 2020) and liquid-frozen (UME RM 2019) RNA-based reference materials in order to be used as a quality control material for SARS-CoV-2 measurements by both clinical laboratories and kit manufacturers.
Materials and methods
Design, cloning, and verification of the fragment containing the SARS-CoV-2 virus genes
The SARS-CoV-2 synthetic RNA fragment (1103 bases) which we designed consists of ten different SARS-CoV-2 target gene fragments and one human internal control gene fragment which were published on protocols released by the World Health Organization (WHO) [16] (Table 1). The Eurofins Genomics synthesized the fragment and cloned it into the pEX-A258 plasmid (Fig. 1). The sequence of clone was confirmed by DNA sequencing and the plasmid was digested with the Hind III restriction enzyme at the 3′ end of the fragment. In order to verify plasmid DNA linearity (3549 bp), the linear plasmid provided by the company was run on a 1% DNA agarose gel and a single DNA band consistent with the DNA length was obtained (data not shown).
Synthesis of SARS-CoV-2 RNA fragment by in vitro transcription
The cloned construct of SARS-CoV-2 was transcribed in vitro using the T7 promoter with 5X MEGAscript™ T7 Kit (Ambion, AMB1334-5) according to the manufacturer’s protocol. Template DNA was removed by TURBO DNase treatment at 37 °C for 15 min. The synthetic RNA was purified using lithium chloride precipitation protocol as stated in the kit.
Quantification and integrity confirmation of in vitro transcribed RNA
The approximate concentration of the synthetic RNA transcript was determined by the Qubit RNA BR assay kit (Quant-iT™). The produced in vitro transcript was diluted to a concentration of about 108 copies/μL. The dilution protocol was given in detail in the “Preparation of RNA reference material” section. The RNA integrity was confirmed with Bioanalyzer 2100 (see Supplementary Information (ESM) Fig. S1).
Verification of RNA transcript by sequence analysis
To confirm the final RNA sequence, in vitro RNA transcript was reverse transcribed, then, multiplied with PCR (Fig. 1) using RdRp-IP2-forward (5′ end of transcript) and RNase P-reverse primer pair (3′ end of transcript) (Table 1). Reverse transcription polymerase chain reaction (RT-PCR) was carried out in a total volume 20 μL using 2x SensiFAST™ Probe No-ROX One-Step Kit (Bioline, BIO-76005). Temperature rates, times, and cycles were as follows: 10 min at 45 °C for reverse transcription, 5 min at 95 °C for polymerase activation, 40 cycles of 15 s at 95 °C for denaturation, and 45 s at 60 °C for annealing and extension. PCR product was purified by High Pure PCR Purification Kit (Roche, 11732676001) according to the manufacturer’s instruction. The amplicon was evaluated and confirmed with Sanger DNA sequencing (Macrogen, Korea) of both strands using primers of RdRp-IP2-forward, ORF1b-Hong Kong-forward, ORF1b-Hong Kong-reverse, and RNase P-reverse. The chromatograms submitted by the company were first checked by eye, subsequently transferred to the Geneious 11.12.1 [21] for editing. The synthetic RNA sequence was deposited in GenBank with the accession number: MW001217.1.
Preparation of RNA reference materials
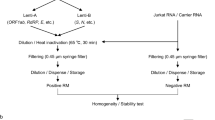
The in vitro transcribed SARS-CoV-2 synthetic RNA was diluted in 1 mM sodium citrate buffer (pH: 6.5, diethylpyrocarbonate treated and autoclaved) containing 2.5 ng/μL total human control RNA (Thermo Fisher 4307281) to an approximate concentration of 108 copies/μL (according to the Qubit RNA BR assay kit (Quant-iT™) measurement) and the tubes were filled in 100 μL volume [22]. All mixing and filling operations were carried out on ice. The RNA reference materials were produced in two different forms; UME RM 2019 was in liquid form and it was kept at −80 °C as frozen for long-term storage, while UME RM 2020, originally filled in 100 μL volume, lyophilized in frozen form, the caps were closed under inert atmosphere and stored at −80 °C. Both forms of reference materials were subjected to characterization, homogeneity, and stability tests required by ISO 17034 [23] and as described in ISO Guide 35 [24].
Homogeneity and stability testing
RNA reference materials were prepared for homogeneity and stability testing as follows: the lyophilized UME RM 2020 were reconstituted by adding 100 μL of ddH2O. UME RM 2019 and UME RM 2020 were both 1000-fold diluted gravimetrically with 10x serial dilutions (100 μL stock + 900 μL ddH2O) in low binding tubes on ice.
Measuring only one gene was enough for homogeneity and stability tests; therefore, we selected one assay and used the measurement results for evaluation of homogeneity and stability. Homogeneity and stability tests were performed targeting the E gene by RT-qPCR, since E gene is one of the most sensitive assays among others [25, 26]. RT-qPCR was carried out in a total volume 20 μL using 2X SensiFAST™ Probe No-ROX One-Step Kit (Bioline) with 5 μL diluted UME RM 2019 or UME RM 2020. Forward primer, reverse primer, and probe were added at a final concentration of 0.9 μM, 0.9 μM, and 0.25 μM, respectively. All the homogeneity and stability tests were performed on LightCycler 480 Real Time PCR instrument (Roche). The reverse transcription was performed at 45 °C for 10 min, following 95 °C enzyme activation for 5 min, and 50 cycles of 95 °C for 15 s and 58 °C for 45 s. For each unit, three PCR replicates were analyzed and the average Cq’s were recorded. All results were generated on the LightCycler 480 v.1.5.62 software with automated settings.
Tubes to be analyzed in homogeneity and stability tests were selected by the Stratified Random Sample Selection Software (TRaNS) developed by TUBITAK UME. A total of 12 tubes of UME RM 2019 and 10 tubes of UME RM 2020 were selected for homogeneity analysis for UME RM 2019 and UME RM 2020, respectively. Short-term stability testing was performed to establish conditions for short-term storage conditions using an isochronous study design [24, 27].
For short-term stability test of UME RM 2019, 2 tubes were exposed to test temperature at −20 °C for 5, 9, 12, and 16 days, while 2 tubes were kept at the reference temperature of −80 °C. For short-term stability test of lyophilized UME RM 2020, 2 tubes were exposed to 4 °C, 25 °C, and 45 °C for 5, 10, 14, and 17 days; as a reference point, 2 tubes were kept at temperature of −80 °C.
For long-term stability test of UME RM 2019, 2 tubes were exposed to test temperature at −20 °C for 1, 2, and 4 months while 2 tubes were kept at the reference temperature of −80 °C. For long-term stability test of lyophilized UME RM 2020, 2 tubes were exposed to three different temperatures −20 °C, 4 °C, and 25 °C for 1, 2, and 4 months. As a reference point, 2 tubes were kept at reference temperature of −80 °C.
Characterization
While UME RM 2019 was characterized using both RT-qPCR and RT-ddPCR, UME RM 2020 was only characterized with RT-qPCR. Characterization of UME RM 2019 and UME RM 2020 by RT-qPCR was performed with 1000-fold 10x serial gravimetric dilutions. RT-qPCR was performed using 3 PCR replicates of three different tubes in 3 different days (a different tube was measured in each day) and two different tubes in two different days for UME RM 2019 and UME RM 2020, respectively. The RT-qPCR conditions were the same as used for homogeneity and stability testing.
RT-ddPCR characterization was only applied to UME RM 2019, which was diluted to about 1000 copy/μL with 10x serial gravimetric dilutions of 3 PCR replicates of two different tubes in 2 different days. The RT-ddPCR conditions were the same as used for homogeneity and stability testing, except the annealing temperature was set to 61 °C for Germany-E assay and China CDC-N assay. RT-ddPCR tests were performed on QX200 (BioRad) platform using double quenched probes (ZEN-IBFQ, IDT) for all gene targets. One-Step RT-ddPCR Advanced Kit for Probes (BioRad, 1864021) was utilized according to the manufacturers’ protocol.
Results
Homogeneity of UME RM 2019 and UME RM 2020
Testing for homogeneity is used to assess if there is a variation between different units of reference materials. Homogeneity analysis of UME RM 2019 and UME RM 2020 was performed using three RT-qPCR replicates. The measurements and regression analyses were performed according to a random sequence to clarify the possible trends in analytical sequence and filling order. No trends in the filling sequence were identified at the 95% and 99% confidence levels for either material. The average Cq values of three measurements for each tested unit were calculated and presented as % in Fig. 2. The relative standard deviation of all samples was calculated as less than 0.4% for both UME RM 2019 and UME RM 2020; thus, it was concluded that both reference materials have sufficient homogeneity suitable for their intended use.
Stability of UME RM 2019 and UME RM 2020
Short-term stability studies were carried out to test the environmental conditions that may occur during the transportation of the material to the user. Reference materials were measured with RT-qPCR and the average Cq values were reported as % average Cq with respect to reference temperature (Fig. 3). No degradation tendency has been observed for both reference materials under the tested conditions; UME RM 2019 was found to be stable for 16 days at −20 °C (Fig. 3a) while the UME RM 2020 was reported to be stable at 4 °C, 25 °C, and 45 °C for 17 days (Fig. 3b).
Long-term stability tests are used to evaluate the stability of materials over a longer period of time for simulating storage conditions in the laboratory. The testing temperatures were selected as −20 °C for UME RM 2019 (Fig. 4a) and − 20 °C, 4 °C, and 25 °C for UME RM 2020 (Fig. 4b). Four months after the production, Cq values of both reference materials showed no significant variations from Cq values of reference temperatures for all tested temperatures (Fig. 4).
Characterization of UME RM 2019 and UME RM 2020
The reference materials UME RM 2019 and UME RM 2020 were both characterized with RT-qPCR (Table 2). For each target gene region, Cq values change between 19.64–24.73 and 19.39–24.69, respectively, depending on the performance of the assays [25, 26]. Similar Cq differences were also observed with in vitro transcribed positive quality control sample of JRC changing from 20.01 to 23.98 [14].
Additionally, UME RM 2019 was only characterized with the RT-ddPCR method (Table 3); it was found that RNA copy numbers changed between 6.1 × 107 and 8.7 × 107 copy/μL in different assays [13, 15]. Similarly, NIST also in vitro produced Research Grade Test Material for SARS-CoV-2 Synthetic RNA Fragments and characterized different gene regions with digital PCR, and mean RNA concentrations of different assays range between 1.9 × 106 and 5.4 × 106 copies/μL copy numbers [13].
Discussion
RNA-based diagnostic techniques play an important role in the diagnosis of SARS-CoV-2 virus with the RT-qPCR method [6]. RNA-based reference materials, UME RM 2019 and lyophilized UME RM 2020, were produced in vitro from a single construct in two different physical forms having ten different SARS-CoV-2 target gene fragments and one human control gene fragment. It is crucial that both enzymatic steps, reverse transcription and PCR amplification, should be included in positive control experiments. RNA-based reference materials are necessary for the development of new measurement methods, and validation of developed methods or kits, as well as comparison of the performance of different diagnostic kits or assays. As UME RM 2019 and UME RM 2020 are RNA-based reference materials, they would ensure covering the activity of both reverse transcription and DNA polymerase enzymes.
Since the produced reference materials contain very high concentration of RNA transcript, about 108 copy/μL, they are suitable for creating calibration graphs with serial dilutions; thus, they would enable comparing the sensitivity of different assays.
For daily internal quality control of RT-qPCR tests and preferentially for the control of each set of PCR, quality control materials of RNA origin should be included. If one tube of this reference material is diluted to 104 RNA copy/μL in a volume of 100 μL, 10,000 tubes of internal quality control material would be produced. Using these reference materials, the quality of the measurement would be ensured for the diagnosis of COVID-19.
The RNA reference materials have been developed in two different forms, liquid frozen and lyophilized. Characterization, homogeneity, and short-term and long-term stability tests were carried out on both forms with RT-qPCR. UME RM 2019 was also characterized with RT-ddPCR. The homogeneity data of UME RM 2019 and UME RM 2020 revealed that the reference materials had sufficient homogeneity for the intended use. While UME RM 2019 can be transported with dry ice, lyophilized UME RM 2020 can be transported to customers at ambient temperature within 2 weeks making it much easier to deliver anywhere in the world. This thermal stabilization approach will greatly facilitate the transfer and the retrieval of the RNA reference materials. UME RM 2019 can be stored at −20 freezer up to 4 months, while UME RM 2020 can be stored at −20 °C, at 4 °C, and even at 25 °C. Once the reference material is diluted, it is recommended to store diluted reference material at −80 freezer and avoid freeze-thaw cycles.
To ensure the quality of the SARS-CoV-2 measurement results, RNA reference materials should be included as positive control material in diagnostic kits and they should also be used as internal quality control in every assay.
References
World Health Organization. Coronavirus Disease 2019 (COVID-19). 2020.
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–9. https://doi.org/10.1038/s41586-020-2008-3.
Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–92. https://doi.org/10.1038/s41579-018-0118-9.
Li C, Yang Y, Ren L. Genetic evolution analysis of 2019 novel coronavirus and coronavirus from other species. Infect Genet Evol. 2020;82:104285. https://doi.org/10.1016/j.meegid.2020.104285.
Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj V V, Guo JZ, Swaney DL. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;1–13.
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045.
Jayamohan H, Lambert CJ, Sant HJ, Jafek A, Patel D, Feng H, et al. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal Bioanal Chem. 2021;413:49–71.
Poon LLM, Chan KH, Wong OK, Yam WC, Yuen KY, Guan Y, et al. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J Clin Virol. 2003;28:233–8.
Benzigar MR, Bhattacharjee R, Baharfar M, Liu G. Current methods for diagnosis of human coronaviruses: pros and cons. Anal Bioanal Chem. 2020.
Jung Y, Park G-S, Moon JH, Ku K, Beak S-H, Lee C-S, et al. Comparative analysis of primer–probe sets for RT-qPCR of COVID-19 causative virus (SARS-CoV-2). ACS Infect Dis. 2020;6:2513–23.
T Nolan, J Huggett, E Sanchez, L (2013). (2013) Good practice guide for the application of quantitative PCR (qPCR), LGC
Sanders R, Mason DJ, Foy CA, Huggett JF. Evaluation of digital PCR for absolute RNA quantification. PLoS One. 2013;8:e75296.
SARS-Cov-2 Synthetic RNA Fragments Guidance Sheet for Research Grade Test Material ID 10169 NIST. https://www.nist.gov/image/sars-cov-2-research-grade-test-material. Accessed 11 Jan 2021.
Synthetic Single Stranded RNA (ssRNA) in Buffer,Product Information Sheet for EURM-019. https://crm.jrc.ec.europa.eu/p/EURM-019. Accessed 11 Jan 2021.
NIM to provide urgently needed 2019-nCoV RNA certified reference materials | National Institute of Metrology of China | NIM. https://en.nim.ac.cn/node/657. Accessed 11 Jan 2021.
World Health Organization. https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pdf. Accessed 11 Jan 2021.
Protocol: real-time RT-PCR assays for the detection of SARS-CoV-2.
Real-time RT-PCR Primers and Probes for COVID-19 | CDC. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. Accessed 11 Jan 2021.
Center for Disease Control and Prevention C. Technical Guidelines for COVID-19 Laboratory Testing. China CDC Wkly. 2020;2:332–6. https://doi.org/10.46234/ccdcw2020.085.
World Health Organisation. https://www.who.int/docs/default-source/coronaviruse/peiris-protocol-16-1-20.pdf?sfvrsn=af1aac73_4. Accessed 11 Jan 2021.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. https://doi.org/10.1093/bioinformatics/bts199.
Fattorini P, Bonin S, Marrubini G, Bertoglio B, Grignani P, Recchia E, et al. Highly degraded RNA can still provide molecular information: an in vitro approach. Electrophoresis. 2020;41:386–93. https://doi.org/10.1002/elps.201900200.
ISO 17034:2016, General requirements for the competence of reference material producers. https://www.iso.org/obp/ui/#iso:std:iso:17034:ed-1:v1:en. Accessed 11 Jan 2021.
ISO Guide 35:2017 - Reference materials -Guidance for characterization and assessment of homogeneity and stability. https://www.iso.org/standard/60281.html. Accessed 11 Jan 2021.
Loan Dao Thi V, Herbst K, Boerner K, Meurer M, Kremer LP, Kirrmaier D, Freistaedter A, Papagiannidis D, Galmozzi C, Stanifer ML, Boulant S, Klein S, Chlanda P, Khalid D, Barreto Miranda I, Schnitzler P, Kräusslich H-G, Knop M, Anders S (2020) A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples.
Vogels C, Brito A, Wyllie A, Fauver J, Ott I, Kalinich C, Petrone M, Landry M, Foxman E, Grubaugh N (2020) Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays. https://doi.org/10.1101/2020.03.30.20048108.
Linsinger TPJ, Pauwels J, Lamberty A, Schimmel HG, Van Der Veen AMH, Siekmann L. Estimating the uncertainty of stability for matrix CRMs. Anal Bioanal Chem. 2001;370:183–8. https://doi.org/10.1007/s002160100719.
Funding
The research was supported by Republic of Turkey East Marmara Development Agency (MARKA) Project No.: TR42/20/COVID/0035.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 207 kb)
Rights and permissions
About this article
Cite this article
Akyurek, S., Demirci, S.N.S., Bayrak, Z. et al. The production and characterization of SARS-CoV-2 RNA reference material. Anal Bioanal Chem 413, 3411–3419 (2021). https://doi.org/10.1007/s00216-021-03284-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03284-w