Abstract
The B.1.1.7 variant of SARS-CoV-2 first detected in the UK harbors amino-acid substitutions and deletions in the spike protein that potentially enhance host angiotensin conversion enzyme 2 (ACE2) receptor binding and viral immune evasion. Here we report cryo-EM structures of the spike protein of B.1.1.7 in the apo and ACE2-bound forms. The apo form showed one or two receptor-binding domains (RBDs) in the open conformation, without populating the fully closed state. All three RBDs were engaged in ACE2 binding. The B.1.1.7-specific A570D mutation introduces a molecular switch that could modulate the opening and closing of the RBD. The N501Y mutation introduces a π–π interaction that enhances RBD binding to ACE2 and abolishes binding of a potent neutralizing antibody (nAb). Cryo-EM also revealed how a cocktail of two nAbs simultaneously bind to all three RBDs, and demonstrated the potency of the nAb cocktail to neutralize different SARS-CoV-2 pseudovirus strains, including B.1.1.7.
Similar content being viewed by others
Main
The emergence of the B.1.1.7 variant of the severe acute respiratory syndrome coronavirus (SARS-CoV-2), also known as the variant of concern (VOC) 202012/01 strain, was first identified in the southeastern United Kingdom (UK) in December 20201. B.1.1.7 was significantly more transmissible than the other SARS-CoV-2 variants across the United Kingdom, with an estimated increase of the reproduction number, also known as R0, by 0.4 to 0.7 (ref. 2). B.1.1.7 quickly spread across the world, resulting in a surge of infected cases. Consequently, stringent travel restrictions and national lockdowns were implemented in many countries. B.1.1.7 is defined by 23 genetic mutations compared to the original strain that was first detected in Wuhan, China. In addition to the aspartic acid (D) to glycine (G) mutation at position 614 (D614G) of the surface spike (S) protein, which has dominated the reported COVID-19 cases since the summer of 2020 (GISIAD database: https://www.gisaid.org), B.1.1.7 harbors eight additional mutations within the S protein. Of particular interest is the asparagine (N) to tyrosine (Y) mutation at position 501 (N501Y), located at the receptor-binding motif (RBM), which is part of the RBD that is involved in direct physical interactions with the human ACE2 to initiate viral entry into host cells3,4,5. While the functional implications of the remaining mutations remain to be established, deletion of histidine 69 and valine 70 (ΔH69/V70) within the N-terminal domain (NTD) of the SARS-CoV-2 S protein is also present in the SARS-CoV-2 mink-associated variant, detected in Denmark, that caused zoonotic transmissions to infect humans, leading to culling of over 17 million minks in November 20206. The proline to histidine mutation at position 681 (P681H) immediately upstream of the polybasic furin cleavage site (Fig. 1a) may affect furin processing of the S protein that is important for membrane fusion and viral entry. Little is known about the potential contributions of the remaining mutations. Although most nAbs that have been identified from convalescent plasma target the RBD of the S protein to sterically prevent its engagement with ACE2, several nAbs were reported to target the NTD without blocking ACE2 binding7,8,9,10,11,12,13. In particular, tyrosine 144 of the S protein, which is deleted in B.1.1.7, is part of the structural epitope of the NTD-specific antibody 4A8. Indeed, a recent study shows that the ΔY144 deletion strongly diminishes binding by multiple NTD-derived nAbs, including 4A8, by more than 1,000-fold, suggesting that the increased infectiousness of B.1.1.7 is associated with its ability to evade host immunity14. We have recently reported a panel of nAbs against the RBD of the SARS-CoV-2 S protein based on mouse hybridoma antibody screening and subsequent engineering into human IgG1 chimeric nAbs15. Among them, three potent nAbs, namely RBD-chAb15, 25 and 45, were found to bind the RBD of the original strain of the SARS-CoV-2 S protein with high potency both in vitro and in vivo. In this study, we examined the impact of B.1.1.7 mutations on the structure and function of the SARS-CoV-2 S protein, and revealed the structural basis of how two nAbs, RBD-chAb15 and 45, simultaneously bind to the RBD of SARS-CoV-2 S protein in a noncompetitive manner to achieve their potent neutralizing activity against SARS-CoV-2 without being compromised by the N501Y mutation.
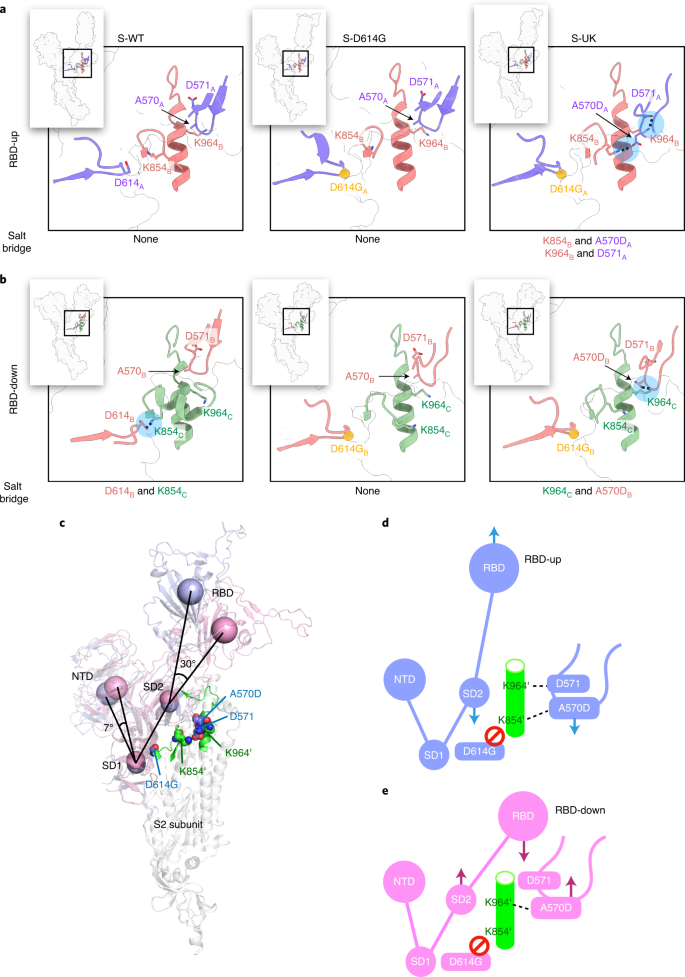
a, Domain structure of S-UK with the nine B.1.1.7 mutations indicated. The D614G mutation common to most emerging SARS-CoV-2 strains is highlighted in blue. b, Four different classes of conformations of S-UK derived from cryo-EM 3DVA. The relative populations of the individual classes are indicated in the pie chart and the number of RBD in the upward conformation (RBD-up) of each class is indicated below, along with the nominal resolution in parentheses. The positions of the RBD-up are indicated by red asterisks. Source data for b are available online (see the source data for Extended Data Fig. 1).
Results
Overall structure of SARS-CoV-2 S-UK (B.1.1.7)
We determined the cryo-EM structure of the ectodomain of the S protein of B.1.1.7 (hereafter S-UK) and identified four distinct conformations by three-dimensional variability analysis (3DVA) (Fig. 1). Three of the conformation classes corresponded to a one upward RBD (RBD-up) conformation (their nominal resolutions ranged between 3.2 and 3.6 Å), and the remaining one corresponded to a two RBD-up conformation with a nominal resolution of 3.3 Å (Fig. 1b, Extended Data Fig. 1 and Table 1). Collectively, the one RBD-up conformations accounted for 73% of the overall population and the two RBD-up conformation accounted for the remaining 27%. Due to the intrinsic dynamics, the loop regions in the NTD where the ΔH69/V70 and ΔY144 lie, as well as the P681H mutation in the furin cleavage site, could not be clearly resolved in the cryo-EM maps. The remaining mutation sites, namely N501Y, A570D, D614G, T716I, S982A and D1118H, could be unambiguously resolved to build the atomic models accordingly (Extended Data Fig. 2). Despite the large number of mutations, the overall structure of S-UK did not differ significantly from that of the original strain detected in Wuhan and the D614G variant (hereafter S-WT and S-D614G, respectively), except for the relative populations and the number of the RBD-up conformation, which have direct implications in host recognition through binding to ACE2 (refs. 4,16,17,18,19,20,21,22).
Regulation of RBD orientation by a pedal-bin mechanism
The RBD-up conformation is the prerequisite for ACE2 binding. Among the reported cryo-EM structures of S-WT solved under physiological conditions (neutral pH), the majority exhibit an all RBD-down state with some in a single RBD-up state18,23,24. In contrast, the reported cryo-EM structures of S-D614G exhibit a higher propensity to populate RBD-up states with the majority being in a one RBD-up state and some in a two RBD-up state19,20,21,22. Our recent study indicated that none of the observed S-D614G particles were in an all RBD-down state, while 62% of which had one RBD-up and 38% had a two RBD-up state25. Similar to S-D614G, S-UK did not populate the all RBD-down state. Nevertheless, relative to S-D614G, S-UK exhibited fewer two RBD-up states (27%) and predominantly populated the one RBD-up state (73%) (Fig. 1b). Gamblin and co-workers postulated that a salt bridge formed by D614 and K854 from two different protomers within the trimeric assembly is a linchpin to restrict interprotomer motions to maintain RBD-down states. The D614G mutation abrogates the D614–K854 salt bridge thereby promoting RBD-up states21,24.
Close examination of the cryo-EM structure of S-UK indicated that the B.1.1.7-specific A570D mutation lies in close proximity to the D614G mutation site. The central helix in which K854 and K964 reside is flanked by D614G and A570D from the other protomer (Fig. 2). The cryo-EM structure of S-UK in an RBD-up state exhibited a new salt bridge formed by K854B and A570DA, accompanied by another salt bridge between K964B and D571A adjacent to A570D (Fig. 2a, bottom panel). In contrast, the cryo-EM structure of S-UK in an RBD-down state exhibited an alternative salt bridge between K964C and A570DB (Fig. 2a, top panel). These additional salt bridges likely compensate for the loss of the salt bridge between D614 and K854 due to the D614G mutation. Comparison of the RBD-up and RBD-down conformations within the same S-UK structure revealed a 30° rotation of the RBD relative to the pivot point defined by the center of mass (COM) of the SD2 domain. In contrast, a minor rotation of the NTD by 7° was observed (Fig. 2b). The rigid-body domain motion resulted in an upward displacement of 22 Å of the COM of RBD with a concomitant downward displacement of 3 Å of the COM of SD2 while the central helix remained static. Note that the downward displacement of SD2 was essentially governed by the downward loop movement promoted by the switch of salt bridge pairing that involves the A570D mutation. We propose that the switching of the A570D-mediated salt bridges may serve as the pedal of a pedal-bin-like mechanism to modulate the RBD motion, which is expected to impact ACE2 binding and viral infectivity. To examine the contribution of the A570D mutation to viral infectivity, we generated a pseudovirus revertant, B.1.1.7+A570, in which the A570D mutation was restored into A570. Compared to B.1.1.7, the revertant was significantly more infectious in the pseudovirus assay (Fig. 3), suggesting that the A570D indeed plays a key role in modulating the RBD conformational transitions in the context of host infection.
a,b, Structural basis of the interprotomer interaction networks around the clinical mutation sites for RBD-up (a) and RBD-down (b). The inset on the upper left corner of each panel illustrates the overall conformation of one protomer with the mutation site indicated by a box, which is expanded in the corresponding panel. The RBD-up and RBD-down protomers are colored in light blue (chain A) and pink (chain B), respectively. For RBD-down, the corresponding protomer, chain B, interacts with another RBD-down protomer, chain C, which is colored in light green. The positions of the salt bridges are highlighted in filled light blue circles. c, Comparison of the relative domain orientations of RBD-up and RBD-down in the context of the clinical mutations. The COMs of NTD (residues 1–288), RBD (residues 330–525), SD2 (residues 317–329 and 526–588) and SD1 (residues 290–316 and 589–695) are shown in spheres over the semitransparent cartoon representations of RBD-up (light blue) and RBD-down (pink). The two mutated residues, D614G and A570D, along with D571 on the same protomer, and K854 and K964 on the other protomer (indicated by an apostrophe), are shown in spheres. The α helix and the preceding loop in which K854’ and K964’ reside are shown in a cartoon representation and colored in green. d,e, Schematic representations of the domain motions of RBD-up (d) and RBD-down (e) modulated by the interprotomer salt bridges. A switch in salt bridge pairing between K964’ and D571/A570D is proposed to regulate the pedal-bin-like motions of RBD and domain D.
HEK293T/hACE2 cells were infected with the B.1.1.7 strain or a revertant B.1.1.7 strain harboring the A570 mutation of SARS-CoV-2 pseudovirus for 72 h. Luciferase activity was measured using a microplate spectrophotometer in two biological replicates, each conducted in three technical replicates. Representative data of two biological replicates are shown. Error bars represent mean values ± s.e.m. A Student’s t-test was used for statistical analysis, which yielded a two-tailed P = 0.0004, t = 11.04 and d.f. = 4. ***P < 0.001.
Molecular basis of ACE2 recognition by S-UK
To glean structural insights into receptor binding of S-UK, we determined the cryo-EM structure of S-UK in complex with the ectodomain of ACE2 fused to a superfold green fluorescence protein (hereafter ACE2). The 3DVA analysis of the complex demonstrated that all three RBDs of S-UK were bound to ACE2 with a 3:3 stoichiometry as opposed to the previously reported heterogeneous binding stoichiometry between the S protein of the variant S-WT and ACE2 (Fig. 4a,b, Extended Data Fig. 3 and Table 2)4,26,27. Furthermore, the local orientation of the RBD in complex with ACE2 appears structurally dynamic, leading to the weak density and poor resolution (>4.5 Å) of this region (Extended Data Fig. 3). We therefore carried out local refinement with a mask focusing on the RBD and ACE2 to generate a density map of 3.5 Å (Extended Data Fig. 3). The improved resolution of the region allowed us to confidently build an atomic model to uncover the binding interface between RBD and ACE2. Detailed inspection revealed a newly formed π–π stacking between the mutated Y501 of S-UK and Y41 of ACE2, which could potentially provide additional stability of the complex formation (Fig. 4c). Furthermore, a contracted rigid-body domain movement of ACE2 towards the RBD of S-UK relative to that of S-WT by 2.5 Å was observed (defined by the COM displacement of ACE2).
a, Orthogonal views of the cryo-EM map of S-UK in complex with ACE2 in a 3:3 binding stoichiometry. b, Expanded view of the atomic model of RBD in complex with ACE2. The RBD of S-UK is colored blue and S-UK-bound ACE2 is colored magenta. For comparison, the RBD of S-WT in complex with ACE2 is shown in gray. The N501Y mutation within the RBD introduced an additional π–π interaction with Y41 of ACE2 that could strengthen their intermolecular interaction. c,d, BLI sensorgrams of S-D614G (c) and S-UK (d) binding to ACE2, which was immobilized on the sensor tip. S-D614G and S-UK concentrations used in independent BLI binding assays are indicated on the right. The kinetic parameters derived from global fitting of the sensorgrams, including the on-rate (kon), off-rate (koff), and dissociation constant (Kd), are shown above, with the fitted data shown in red lines that are overlaid with the experimental curves. Source data for a and b are available online (see the source data for Extended Data Fig. 3).
To evaluate the impact of the B.1.1.7-specific mutations on the binding of ACE2, we immobilized ACE2 onto the sensor tips for biolayer interferometry (BLI) and compared the binding kinetics to S-UK and S-D614G. A 2.4-fold decrease in the dissociation constant (Kd) of S-UK (Kd = 1.3 nM) compared to that of the D614G variant (Kd = 3.1 nM) was observed (Fig. 4d). The main difference was the reduced off-rate of S-UK (1.1 × 10−4 s−1) compared to S-D614G (5.3 × 10−5 s−1). The increased binding affinity of S-UK for ACE2 was consistent with the previous report in which the N501Y mutation was introduced to the unmutated S protein; in that case, the Kd value for ACE2 binding was modestly reduced from 8.6 nM to 6.3 nM based on similar BLI measurements5.
The N501Y mutation helps evade antibody neutralization
In addition to enhancing ACE2 binding, the mutations in S-UK could help SARS-CoV-2 evade neutralizing antibodies to achieve higher infectivity14. To address this possibility, we tested the abilities of three chimeric anti-RBD antibodies (RBD-chAbs) to compete with ACE2 binding to S-UK. We recently demonstrated that the three RBD-chAbs, namely RBD-chAb15, 25 and 45, targeted three distinct sites within the RBM, that each of these RBD-chAbs potently neutralize pseudovirus of the original Wuhan strain of SARS-CoV-2, and that a cocktail of RBD-chAb25 and 45 can prophylactically protect mice and hamsters from SARS-CoV-2 infection15. BLI analysis showed that pre-incubation of S-D614G with each of the three RBD-chAbs effectively prevented ACE2 binding. However, RBD-chAb25 failed to compete for ACE2 binding to S-UK, while RBD-chAb15 and 45 remained highly effective ACE2 binding inhibitors (Fig. 5a–c). The loss of neutralizing activity of RBD-chAb25 against S-UK can be rationalized by the overlap of its structural epitope and the N501Y mutation revealed by our cryo-EM analysis (Fig. 4c, Extended Data Fig. 4 and Table 2). Instead, epitopes targeted by RBD-chAb15 and 45 reside on two distal ends of RBM, thus potentially allowing simultaneous binding to the same RBD. To verify our hypothesis, we sequentially mixed RBD-chAb45 and 15 with S-D614G shortly before cryo-EM grid vitrification, and determined the cryo-EM structure of the quaternary complex (S-D614G:RBD-chAb15/45). The resulting cryo-EM map revealed stoichiometric binding of RBD-chAb15 and 45 to all three RBDs, and the N501 was bypassed by the two RBD-chAbs (Fig. 5). Despite the limited space above the three upward pointing RBDs to accommodate six antibodies, the poses of RBD binding by the two RBDs were essentially identical to that of individually bound cryo-EM structures, underscoring their potential to be used as a cocktail therapy for B.1.1.7.
a–c, BLI sensorgrams of immobilized ACE2 binding to S-D614G (yellow) and S-UK (indigo) that were preincubated with RBD-chAb15 (a), RBD-chAb25 (b) and RBD-chAb45 (c). Only S-UK preincubated with RBD-chAb25 showed significant ACE2 binding, reflecting loss of neutralizing activity of RBD-chAb25 due to the N501Y mutation. The gray arrow indicates the time point at which complex dissociation was triggered. d, Orthogonal views of the cryo-EM map of S-D614G in complex with RBD-chAb15 and 45 with 3:3:3 binding stoichiometry. The three RBDs are shown in blue; heavy and light chains of RBD-chAb15 are shown in amber and yellow, respectively; heavy and light chains of RBD-chAb45 in forest green and apple green, respectively. e, Superposition of cryo-EM structure of RBD-bound ACE2 (PDB 7KMS)4 and the quaternary complex of the antibody cocktail and RBD. The N501Y mutation site is shown in a red sphere. f, Structural mapping of the ACE2 binding interface, defined as atoms within RBD that are within 5 Å of ACE2, is colored in magenta. g, Structural mapping of the antibody cocktail binding interface. The RBD-chAb15 binding interface is colored in green and the RBD-chAb45 binding interface is in orange. h, Structural mapping of clinically reported mutations within RBD associated with the VOCs B.1.1.7, B.1.351, P1 and B.1.617. The positions of the mutations are colored in orange. N501Y, the only RBD mutation of S-UK, is indicated by an asterisk. i, Structural mapping of the contacting frequency of nAbs derived from convalescent sera. In total, 46 cryo-EM or crystal structures of nAb-bound RBD were analyzed by PISA to define the intermolecular contacts. The frequency of nAb contacts is indicated in the heat map on the right. The identities of the most targeted residues are indicated. Source data for d are available online (see the source data for Extended Data Fig. 4).
To further verify our hypothesis, we generated two pseudoviruses that individually expressed S-D614G and S-UK on the viral surface, and tested the neutralization activity of RBD-chAb15 and 45 individually and in combination (cocktail). The results showed that RBD-chAb45 was more effective than RBD-chAb15, that both nAbs were more effective against the D614G strain (Fig. 6a) over the B.1.1.7 strain (Fig. 6b), and that the cocktail of equal amounts of RBD-chAb15 and 45 was significantly more effective than the separate nAbs. The effect of the antibody cocktail appeared to be additive when the three datasets were global fit to the Bliss independence model28.
a,b, The neutralization effects of RBD-chAb15, 45 and their combination against pseudovirus strain D614G (a) and strain B.1.1.7 (b). Assays were carried out in triplicate and plotted individually. The results are a global fit to an additive Bliss independent model by sharing the half-maximum inhibitory concentration (IC50) values of RBD-chAb15 and 45 (Methods). The best-fit IC50 values and the corresponding 95% confidence ranges (shown in parentheses) of RBD-chAb15 and 45, and their combination derived from a and b are tabulated on the right. aThe IC50 values of the antibody cocktails RBD-chAb15/45 against D614G and B.1.1.7 when the titration curves are analyzed separately (the corresponding 95% confidence ranges are shown in parentheses below).
Discussion
In summary, we showed that the B.1.1.7-specific mutations minimally perturb the overall structure of the trimeric assembly of S-UK while a shift in the population of the RBD-up state was observed. Compared to S-D614G, the two RBD-up population of S-UK was reduced without populating the all RBD-down state. Cai et al. recently reported a similar trend, observing that the S-UK showed increased one RBD-up population (82%) while a reduced population (15%) of the all RBD-down state could still be identified, and only a small fraction (3%) was in the two RBD-up state29. On the other hand, Gobeil et al. found three-quarters of the S-UK in the one RBD-up state, and the other quarter in the all RBD-down state30. We identified a unique salt bridge switch involving the B.1.1.7-specific A570D mutation with two lysine residues, K854 and K964, in the central helix that compensate for the loss of the salt bridge between D614 and K854 due to the D614G mutation. We postulate that the newly evolved A570D mutation serves as a molecular switch in a pedal-bin mechanism to kinetically modulate the up/down motion of RBD. The RBD-up state is stabilized by the double salt bridges, A570D–K854 and D571–K964, while the RBD-down state is stabilized by the A570D–K964 salt bridge. The latter resembles that of the original D614–K854 salt bridge in S-WT (Fig. 2). The new salt bridge formed between A570D and K854 from the other protomer was also reported by Cai et al. in the one RBD-up state29. Additionally, both Cai et al. and Gobeil et al. reported that A570D forms an interprotomer hydrogen bond with N856 from another protomer in the RBD-down state30, whereas we observed A570D to form a salt bridge with K964 from a different protomer, suggesting some degree of conformational heterogeneity in this state (Extended Data Fig. 5). Although the RBD-up and -down populations of S-UK varied between different studies, and the interprotomer interactions associated with A570D also differed in some cases, a converging view is that the A570D mutation may play an important role in modulating the RBD conformation. Indeed, we showed by the pseudovirus assay with a pseudovirus revertant, B.1.1.7+A570, that the A570D mutation makes significant contributions to modulate the infectivity (Fig. 3).
The fact that our S-UK:ACE2 structure adopts a 3:3 binding stoichiometry while the majority of the previously reported cryo-EM structures of ACE2-bound S-WT display a single RBD-up engaged with ACE2 binding4,26,27 suggests that the additional double salt bridges help kinetically stabilize the RBD-up state in S-UK. Note that natural ACE2 is dimeric as opposed to the monomeric ectodomain of ACE2 used in our study. The exact binding stoichiometry and affinity between ACE2 and the S protein variants may vary. Nevertheless, Cai et al. used an Fc-fused ACE2, which exists as a dimer, to assess the S protein binding. In their case, S-UK was found to bind ACE2-Fc with a fivefold stronger affinity than S-D614G29, in line with our findings. Furthermore, our S-UK harbors tandem proline mutations, the furin cleavage site mutations and the C-terminal foldon sequence for stabilization purposes, which may operate differently in the natural spike context.
According to our cryo-EM structure of S-UK, the N501Y mutation in S-UK introduces π–π staking between N501Y and Y41 of ACE2. The N501Y mutation is most likely responsible for the 2.4-fold increase of binding affinity between S-UK and ACE2 compared to that between S-D614G and ACE2, according to our BLI measurements (Fig. 4). A similar finding has been reported by Subramanian and colleagues when the N501Y mutation was introduced to the original S spike without the other B.1.1.7-specific mutations, including the prevalent D614G mutation5. In the context of antibody neutralization, S-UK harbors the N501Y mutation within RBM to disrupt the neutralizing effects of several potent antibodies14, including RBD-chAb25 in our case. Nevertheless, RBD-chAb15 and 45 remained highly neutralizing in the context of competing ACE2 binding (Fig. 5a–g). Subramanian and colleagues also reported two potently neutralizing antibodies that target the RBD with the N501Y mutation5. The structural epitopes of the two antibodies, VH ab8 and Fab ab1, partially overlap. While VH ab8 targets the RBD-down conformation, Fab ab1 targets the RBD-up conformation. In this study, the quaternary cryo-EM structure of S-D614G:RBD-chAb15/45 showed that the two antibodies could simultaneously bind to the same RBD without contacting the N501Y mutation site. While the epitope of RBD-chAb45 is similar to that of VH ab8, the epitope of RBD-chAb15 does not overlap with that of VH ab8 or Fab ab1 (Extended Data Fig. 6). The pseudovirus assay further demonstrated that the combined use of RBD-chAb15 and 45 can additively neutralize pseudoviruses that correspond to D614G and B.1.1.7 (Fig. 6).
RBD-chAb45 targets the tip of the RBM where F486 resides. Statistics of reported SARS-CoV-2 S protein bound to convalescent plasma-derived nAbs indicated that F486 is one of the most targeted RBD residues (Fig. 5i). The adjacent E484K mutation that is present in the emerging South African 501.v2 strain (B.1.351) and the Brazilian P1 strain could potentially reduce the efficacy of RBD-chAb45. Additional point mutations are identified in several VOCs31, including B.1.351 and P1, which partially overlap with the high-frequency nAb epitopes and potentially impede their neutralizing activities (Fig. 5h,i). For instance, combinations of E484K and N501Y are present in the B.1.351 and P1 variants, which harbor additional K417N and K417T mutations, respectively. E484K together with several B.1.351-specific mutations within the RBD have been shown to be refractory to several RBD-targeting nAbs14. L452R that is present in the variant B.1.617 (first detected in India), the variant B.1.427 (first detected in California) and the variant B.1.526.1 (first detected in New York) also renders some nAbs less effective. Nevertheless, the structural epitope of RBD-chAb15 is less frequently recognized by convalescent plasma-derived nAbs, and there is hitherto no mutation in the more transmissible SARS-CoV-2 variants that are located within the epitope of RBD-chAb15 (Fig. 5g). Therefore, the ability of RBD-chAb15 and 45 to simultaneously bind to distinct regions of RBD is an attractive feature when considering their use in prophylactic cocktail antibody therapy to prevent mutational viral escape.
Methods
Expression and purification of S-UK and S-D614G
The codon-optimized nucleotide sequence of S-UK harboring the double proline mutations (2 P, 986KV987 → 986PP987) and the furin cleavage site mutation (fm, 682RRAR685 → 682GSAG685) for stabilizing the S protein in a prefusion state32 was a gift of M.-H. Tao (Institute of Biomedical Sciences, Academia Sinica). The DNA sequence corresponding the residues 1–1208 of S-UK was subcloned into the mammalian expression vector pcDNA3.4-TOPO (Invitrogen), which contains a foldon trimerization domain based on phage T4 fibritin followed by a c-myc epitope and a hexa-repeat histidine tag as described previously33. The same construct design was used for S-D614G as described elsewhere25.
S-D614G and S-UK plasmids were transiently transfected into HEK293 Freestyle cells for recombinant protein production as described previously33. The recombinant proteins were affinity purified by overnight binding to HisPur Cobalt Resin (Thermo Fisher Scientific) in buffer A (50 mM Tris-HCl (pH 7.6), 300 mM NaCl, 5 mM imidazole and 0.02% NaN3 at 4 °C), followed by washing with buffer B (50 mM Tris-HCl (pH 7.6), 300 mM NaCl, 10 mM imidazole) and elution by buffer C (50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 150 mM imidazole). The eluted protein was further purified by size-exclusion chromatography (SEC) using a size-exclusion column (Superose 6 increase 10/300 GL; GE Healthcare) in TN buffer (50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 0.02% NaN3). The protein concentrations were determined by using the UV absorbance at 280 nm using an UV–Vis spectrometer (Nano-photometer N60, IMPLEN).
Expression and purification of sfGFP-ACE2
The sfGFP-ACE2 construct was obtained from Addgene (Plasmid No. 145171), which was deposited by E. Proco (University of Illinois, USA)34. Recombinant sfGFP-ACE2 was transiently expressed in Expi293 cell and secreted into the culture medium. sfGFP-ACE2 was purified by an ion-exchange column (HiTrap 5 ml Q FF anion-exchange chromatography column; GE Healthcare) eluted by a linear salt gradient from 10 to 1000 mM NaCl, followed by SEC using a size-exclusion column (Superdex 75 16/600 GL; GE Healthcare) in TN buffer. The protein concentrations were determined by using the UV absorbance at 280 nm using a UV–Vis spectrometer (Nano-photometer N60, IMPLEN).
Preparation of S-UK in complex with sfGFP-ACE2
Three microliters of S-UK in 2.3 mg ml−1 was mixed with 1.2 equivalent of sfGFP-ACE2 (S-UK:sfGFP-ACE2) and incubated at room temperature for 1 h before being separated by SEC using a size-exclusion column (Superose 6 increase 10/300 GL; GE Healthcare) in TN buffer. The elution peak corresponding to S-UK in complex with sfGFP-ACE2 was collected and concentrated to 1.5 mg ml−1 for cryo-EM grid preparation.
Preparation of S-D614G in complex with RBD-chAb cocktail
Three microliters of S-D614G in 1.2 mg ml−1 were mixed with 1.2 equivalent of RBD-chAb45 and incubated at room temperature for 1 h before being separated by SEC using a size-exclusion column (Superose 6 increase 10/300 GL; GE Healthcare) in TN buffer. The elution peak corresponding to S-UK in complex with RBD-chAb45 was collected and concentrated to a total concentration of 1.5 mg ml−1, followed by the addition of 1.2 equivalent RBD-chAb15 (S-UK:RBD-chAb15/45) for cryo-EM grid preparation. The preparation of RBD-chAb15 and 45 was described elsewhere15.
Cryo-EM sample preparation and data collection
Three microliters of purified protein samples—S-UK, S-UK:sfGFP-ACE2 and S-D614G:RBD-chAb15/45—were applied onto 300-mesh Quantifoil R1.2/1.3 holey carbon grids. The grids were glow-charged at 20 mA for 30 s. After 30 s incubation, the grids were blotted for 2.5 s under 4 °C with 100% humidity and vitrified using a Vitrobot Mark IV (Thermo Fisher Scientific). Data acquisition was performed on a 300 keV Titan Krios microscope equipped with a Gatan K3 direct detector (Gatan) in the super-resolution mode using EPU 2.10 software (Thermo Fisher Scientific). Videos were collected with a defocus range of −0.8 to −2.6 µm at a magnification of ×81,000, resulting in a pixel size of 0.55 Å. A total dose of 50 e−/Å2 was distributed over 50 frames with an exposure time of 1.8 s. The dataset was collected with an energy filter (slit width 15–20 eV) and the dose rate was adjusted to 8 e− pixel−1 s−1.
Image processing and 3D reconstruction
All ×2 binned super-resolution raw videos were subject to Relion v.3.0 (ref. 35) with dose-weighting and 5 × 5 patch-based alignment using GPU-based software MotionCor v.2 (ref. 36). After motion correction, all corrected micrographs were then transferred to cryoSPARC v.3.0.1 (ref. 37). Contrast transfer function (CTF) estimation was performed by patch-based CTF. The satisfied exposures, whose ‘CTF_fit_to_Res’ parameters were between 2.5 and 4 Å, were selected and applied to particle picking. A small subset of micrographs was used for the template-free blob picker. The picked particles were extracted with a box size of 384 pixels and followed by iterative rounds of two-dimensional (2D) classification for filtering junk particles.
For S-UK, approximately 1.3 million particles were picked and classified by ab-initio reconstruction with C1 symmetry (class = 5), followed by heterogeneous refinement to generate five distinct classes (class = 5) and non-uniform refinement to generate an initial model. A mask was generated based on the initial 3D model for 3DVA by cryoSPARC v3.0.1 to generate five clusters of structural ensembles. One of the five clusters, which accounted for 16% of the initial particle images, yielded a poor initial 3D model that limited further refinement to define the conformations of the three RBDs. We therefore focused on the four remaining classes for further refinements. Particle images corresponding to four of the five clusters were re-extracted, un-binned and non-uniformly refined with local CTF refinement to yield four cryo-EM maps with nominal resolutions ranging between 3.1 and 3.6 Å (Extended Data Fig. 1).
A similar imaging processing workflow was applied for S-UK:sfGFP-ACE2 and S-D614G:RBD-chAb15/45. For S-UK:sfGFP-ACE2, five clusters of structural ensembles were also generated by 3DVA, but all particle images (n = 463,191) were pooled and refined without symmetry constrain to yield a 3.3-Å cryo-EM map (Extended Data Fig. 3). The local resolution analysis was calculated using ResMap38. To improve the local resolution of the ACE2 binding interface, a mask was created to include ACE2 and RBD for local refinement, which yielded a 3.5-Å cryo-EM of the subset of the S-UK:sfGFP-ACE2 structure. For S-D614G:RBD-chAb15/45, approximately 2.3 million particles were picked for iterative 2D classification, which yielded 78,790 particles for initial model building, and templated particle picking, which generated 108,126 particles for 3D classification into three classes. The largest class (74%) was further refined by using a C3 symmetry to yield a 4.0-Å cryo-EM map (Extended Data Fig. 4). The local resolution analysis was calculated using ResMap38.
Model building and refinement
An initial model of S-UK was generated based on PDB 6XM3 by using Swiss-Model39. The coordinate was divided into individual domains and manually fitted into the cryo-EM map in UCSF-Chimera v.1.14 (ref. 40), UCSF-ChimeraX v.1.1.1 (ref. 41) and Coot v. 0.9.3 (ref. 42). A similar approach was used for S-UK:sfGFP-ACE2, for which the crystal structure of ACE2 in complex with RBD (PDB 7KMS) was used as the initial model. For S-UK:RBD-chAb15/45, the previously determined structures of S-WT in complex with RBD-chAb15 and RBD-chAb45 were used as a template for model building15. After iterative refinements, the coordinate was further processed by real-space refinement in Phenix v.1.18 (ref. 43) to obtain a convergent model. Based on the role of the N-glycosylation motif (N-X-S/T), N-linked glycans were built onto asparagine side-chains by using the extension module ‘Glyco’ within Coot42. The final models were assessed by MolProbity44. Structural visualization and rendering of structural representations were achieved by using a combination of UCSF-Chimera v.1.14, UCSF-ChimeraX v. and Pymol v.2.4.0 (Schrodinger Inc.).
Biolayer interferometry
A stock solution of sfGFP-ACE2 was diluted to at 10 µg ml−1 for immobilization onto High Precision Streptavidin (SAX) biosensors (Sartorius) in assay buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.02% NaN3, 0.1% BSA) according to manufacturer’s instructions. Stock solutions of S-D614G and S-UK were serially diluted to 100, 50, 25, 12.5, 6.25 and 3.125 nM in assay buffer for independent binding using an OctetRED 96 biolayer interferometer (ForteBio). The association and dissociation were monitored over 600 s and 900 s, respectively. The six independent BLI sensorgrams with different S protein concentrations were baseline corrected using double reference, that is, biosensors without sfGFP-ACE2 were used to collect sensorgrams with another set of S protein samples to minimize nonspecific binding and to extract the actual responses from the protein-protein interactions. The double reference subtracted data were globally fit to a 1:1 binding model using Data Analysis v.10.0 software (ForteBio). For RBD-chAb neutralization analysis, RBD-chAb15, 25 and 45 were individually mixed with an equal molar ratio of S-D614G or S-UK at a final concentration of 100 nM, and incubated at room temperature before BLI measurements using the same sfGFP-ACE2-immobilized biosensors with 400 s of association and 150 s of dissociation. The results were exported and replotted Prism v.9 (GraphPad).
Structure-based statistics of RBD epitope binding frequency
The identification of structural epitopes of convalescent sera-derived RBD-specific nAbs at a residue-specific level were achieved by using the ‘Protein interfaces, surfaces and assemblies’ service PISA at the European Bioinformatics Institute (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html)45. The list of cryo-EM and crystal structures of nAb-bound spike and RBD was manually curated by searching the entries deposited in the Protein Data Bank and their corresponding publications as tabulated in Supplementary Tables 1 and 2. The nAb contacting residues within RBD were defined by the function ‘Interfaces’ of PISA and summarized in Supplementary Table 3. In the cases where one RBD residue is in contact with both the heavy chain and a light chain of a given nAb, the epitope count was considered twice. The cumulated epitope counts were mapped onto the crystal structure of RBD in complex with ACE2 (PDB 6M0J) and rendered by using Pymol v.2.4.0 (Schrodinger Inc.).
Pseudovirus infection and antibody neutralization assay
The pseudovirus infection and antibody neutralization assays were performed using HEK293T cells overexpressing human ACE2 (HEK293T/hACE2) and SARS-CoV-2 pseudotyped lentivirus expressing full-length S protein provided by the National RNAi Core Facility (Academia Sinica, Taiwan). Into each well of a 96-well white plate (Corning Costar), 1 × 104 HEK293T/hACE2 were seeded and cultivated for 16 h at 37 °C. Fourfold serially dilution of chAbs were premixed with 1,000 TU per well of D614G and B.1.1.7 strains of SARS-CoV-2 pseudovirus. The mixture was incubated for 1 h at 37 °C and then added to pre-seeded HEK293T-ACE2 cells at 100 μl per well for 24 h at 37 °C. In the case of the B.1.1.7+A570 revertant infection assay, the B.1.1.7 strain was used as a control and two independent experiments were carried out, each with three technical replicates for statistical analyses. The pseudovirus-containing culture medium was removed and replaced with 100 μl per well of DMEM for additional 48-h incubation. Next, 100 μl of ONE-Glo luciferase reagent (Promega) was added to each well for 3-min incubation at 37 °C. The luciferase activities were measured using a microplate spectrophotometer (Molecular Devices). The observed luciferase activities were normalized by the values derived from DMEM with and without pseudovirus in the absence of antibodies, which were defined as 0% and 100% inhibition, respectively. The observed inhibition rate was plotted as a function of antibody concentration and was calculated by comparing the luminescence value to the negative and positive control wells. The Bliss independence model28 was evoked to evaluate whether the system was additive or not. The three experimental titration curves of antibody neutralization were fit to the following equations:
where Ya and Yb are normalized inhibition when the two antibodies, RBD-chAb15 and 45, were tested separately. Yab is the normalized inhibition when the two antibodies are combined in a 1:1 ratio. X is the logarithm of antibody concentration and n is the Hill’s coefficient, which is assumed to be the same for both antibodies. Ya, Yb and Yab were globally fit to the equations above by sharing the IC50,a and IC50,b and the Hill’s coefficient. If there were clear deviations between the experimental data points and the global fitting results, it would suggest that the additive model is inadequate. GraphPad Prism v.9 (GraphPad Software Inc.) was used for the global fitting procedure and the independent IC50 fitting procedure.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Atomic coordinates of S-UK are deposited in the Protein Data Bank (PDB) under the accession codes 7EDF, 7EDG, 7EDH and 7EDI. Those of S-UK:ACE2 and S-D614G:RBD-chAb15/45 are deposited in the PDB under the accession codes 7EDJ and 7EH5, respectively. Cryo-EM maps are deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-31069 to EMD-31074. Source data are provided with this paper.
References
Galloway, S. E. et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage – United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal. Wkly Rep. 70, 95–99 (2021).
Volz, E. et al. Transmission of SARS-CoV-2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. Preprint at https://doi.org/10.1101/2020.12.30.20249034 (2021).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280 e8 (2020).
Zhou, T. et al. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe 28, 867–879 e5 (2020).
Zhu, X. et al. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. 19, e3001237 (2021).
Oude Munnink, B. B. et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371, 172–177 (2021).
Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020).
Greaney, A. J. et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29, 44–57 e9 (2021).
Barnes, C. O. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020).
Chi, X. et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369, 650–655 (2020).
Liu, L. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020).
Lv, Z. et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 369, 1505–1509 (2020).
Tortorici, M. A. et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957 (2020).
Wang, P. et al. Antibody resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021).
Su, S. C. et al. Structure-guided antibody cocktail for prevention and treatment of COVID-19. PLoS Pathogens (in the press).
Shang, J. et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020).
Yan, R. et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 (2020).
Walls, A. C. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292 e6 (2020).
Yurkovetskiy, L. et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183, 739–751 (2020).
Gobeil, S. M. et al. D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 34, 108630 (2021).
Benton, D. J. et al. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc. Natl Acad. Sci. USA 118, 33579792 (2021).
Zhang, J. et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 372, 525–530 (2021).
Hsieh, C. L. et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020).
Wrobel, A. G. et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 27, 763–767 (2020).
Yang, T. J. et al. COVID-19 dominant D614G mutation in the SARS-CoV-2 spike protein desensitizes its temperature-dependent denaturation. Preprint at https://www.biorxiv.org/content/10.1101/2021.03.28.437426v1 (2021).
Xu, C. et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 7, eabe5575 (2021).
Xiao, T. et al. A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent. Nat. Struct. Mol. Biol. 28, 202–209 (2021).
Bliss, C. I. The toxicity of poisons applied jointly. Ann. Appl. Biol. 26, 585–615 (1939).
Cai, Y. et al. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. Science https://doi.org/10.1126/science.abi9745 (2021).
Gobeil, S. M. et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science https://doi.org/10.1126/science.abi6226 (2021).
Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions; https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html (2021).
Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020).
Yang, T. J. et al. Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans. Proc. Natl Acad. Sci. USA 117, 1438–1446 (2020).
Chan, K. K. et al. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 369, 1261–1265 (2020).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42766 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
We thank the Academia Sinica Biophysics Core Facility (grant no. AS-CFII108-111) and the Academia Sinica Cryo-EM Center (grant no. AS-CFII-108-110) for data collection, and the Academia Sinica RNAi Core Facility for providing SARS-CoV-2 pseudotyped lentiviruses, all of which are funded by the Academia Sinica Core Facility and Innovative Instrument Project. We also thank the mammalian cell culture facility of the Institute of Biological Chemistry, Academia Sinica, for supporting protein production and the Academia Sinica Grid Computing for cryo-EM data processing. This research was supported by the funding supports from Academia Sinica to H.-C.W. (grant no. AS-CFII-108-102) of IBMS P3 facility, an Academia Sinica Career Development Award (grant no. AS-CDA-109-L08) and an Infectious Disease Research Supporting Grant (grant no. AS-IDR-110-08) to S.-T.D.H., and the Ministry of Science and Technology, Taiwan (grant nos. MOST-108-3114-Y-001-002 and MOST-108-2823-8-001-001 to H.-C.W, and MOST 109-3114-Y-001-001 to H.-C.W. and S.-T.D.H.).
Author information
Authors and Affiliations
Contributions
S.-T.D.H. and H.-C.W. conceived the experiment. T.-J.Y. and P.-Y.Y. prepared the spike proteins and ACE2. W.-Y.C and H.-T.L. generated and prepared RBD-chAbs. T.-J.Y. and Y.-C.C. collected the cryo-EM data. T.-J.Y. processed the cryo-EM data and modeled the structures. T.-J.Y. and M.-R.H. collected and analyzed BLI data. K.-H.L and H.-C.T. collected and analyzed pseudovirus data. P.-Y.Y. curated the RBD epitope usage data. T.-J.Y. and S.-T.D.H. wrote the manuscript with inputs from all co-authors. S.-T.D.H. and H.-C.W. obtained funding.
Corresponding author
Ethics declarations
Competing interests
Related to this work, the Academia Sinica has filed a US patent application for the neutralizing antibodies on which H.-C.W. and S.-T.D.H. are named as inventors. The patent application is pending approval. The other authors declare no competing interests.
Additional information
Peer review information Nature Structural and Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Workflow for cryo-EM data processing of apo S-UK.
Overview of the cryo-EM data processing of apo S-UK. The micrographs that meet the selection criterion of CTF_fit_resolution between 2.5 and 4 Å were used for particles selection, multiple rounds of 2D averaging classification, initial 3D classification, and non-uniform (NU) refinement. The resulting initial map and corresponding mask from the NU-refinement were used for 3D variability analysis (3DVA) followed by another round of heterogeneous refinement. Four classes of structures were resolved from the dataset, which yielded three distinct structures in the one RBD-up state, and one structure in the two RBD-up state. A representative micrograph as the source data is available online (Source Data Fig. 1).
Extended Data Fig. 2 Expanded views of cryo-EM maps and atomic models of apo S-UK at the UK variant-specific mutations sites.
The individual expanded views are taken from the one RBD-up conformation 1 (see Supplementary Table 1).
Extended Data Fig. 3 Workflow for cryo-EM data processing of S-UK:ACE2.
Overview of the cryo-EM data processing of SARS-CoV-2 S-UK in complex with the ACE2 ectodomain. The same processing procedure as described in Extended Data Fig. 1 was used to yield five distinct structures based on the 3DVA. Each structure is shown in semitransparent surface and fit to the most populated structure (solid pink structure) from the top view to highlight the domain movements of the individual ACE2 ectodomains. To obtain a high-resolution structure of ACE2 in complex with the S-UK, a mask was generated (semitransparent volume in the bottom gray EM map) for the local refinement, which yielded a 3.5-Å cryo-EM map as shown on the bottom right panel. ACE2 and RBD are colored in magenta and blue, respectively. A representative micrograph as the source data is available online (Source Data Fig. 2).
Extended Data Fig. 4 Workflow for cryo-EM data processing of S-D614G:RBD-chAb15/45.
Overview of the cryo-EM data processing of SARS-CoV-2 S-D614G in complex with RBD-chAb15 and RBD-chAb45. The same processing procedure as described in Extended Data Fig. 1 was used to yield three distinct structures based on the 3DVA. The selected class (gray) was used for the NU-refinement to yield a 4.0-Å cryo-EM map. The local resolutions are mapped onto the EM map with the scale bar shown on the right. A representative micrograph as the source data is available online (Source Data Fig. 3).
Extended Data Fig. 5 A570D-associated inter-protomer interactions within S-UK.
The expanded views of the reported S-UK structures superimposed with the current study. Top panels. S-UK structures in the RBD-up state. The S-UK structure reported in this study is colored in tomato. Bottom panels. S-UK structures in the RBD-down state. The S-UK structure reported in this study is colored in orchid. The S-UK structures reported by Cai et al.1 (PDB entry 7N1Y) and Gobeil et al.2 (PDB entry 7LWV) are shown in semitransparent gray and magenta, respectively. The corresponding protomer chain IDs are indicated in subscripts. In the RBD-up state, Cai et al. observed the same inter-protomer salt bridge between A570D and K854 while the corresponding K854 is missing in the atomic model reported by Gobeil et al. In the RBD-down state, both Cai et al. and Gobeil et al. reported an inter-protomer hydrogen bond between A570D and N856 whereas we observed an inter-protomer salt bridge between A570D and K964 from the other protomer opposite of the other studies’ findings. The inter-protomer salt bridges and hydrogen bonds are indicated by black dashed lines.
Extended Data Fig. 6 Comparison of antibody epitopes of our current study and previously reported antibodies against the S protein harboring the N501Y mutation at RBD.
Left panel. RBD is shown in surface representation with the residues that are in contact with the light chain and heavy chain of RBD-chAb15 (Ab15) colored in lime and forest green, respectively. The residues that are in contact with the light chain and heavy chain of RBD-chAb45 (Ab45) colored in light yellow and gold, respectively. Middle panel. RBD is shown in surface representation with the residues that are in contact with the VH ab8 (PDB ID: 7MJI) colored in steel blue. Right panel. RBD is shown in surface representation with the residues that are in contact with the light chain and heavy chain of Fab ab1 (PDB ID: 7MJL) colored in light blue and steel blue, respectively. In all cases, N501Y is color in red to highlight how these antibodies bypass the mutation to retain their binding to RBD. The number of RBD-up or RBD-down conformation within the trimeric S protein structure is indicated within parentheses below. In our structure, two nAbs binding simultaneously to the same RBD while in the work by Subramanian and colleagues, the two antibodies bind separately the one RBD3.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2.
Supplementary Table 3
Residue-specific epitope mapping of RBD-directed nAbs. The contacts are defined by EMBL-EBI. The residues that are in contact with either the heavy or light chain are highlighted in pink. The residues that are in contact with both the heavy and light chains are highlighted in brick red.
Source data
Source Data Extended Data Fig. 1
Representative micrograph of apo-S-UK.
Source Data Extended Data Fig. 3
Representative micrograph of S-UK in complex with ACE2.
Source Data Extended Data Fig. 4
Representative micrograph of S-UK in complex with RBD-chAb15 and 45.
Rights and permissions
About this article
Cite this article
Yang, TJ., Yu, PY., Chang, YC. et al. Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function. Nat Struct Mol Biol 28, 731–739 (2021). https://doi.org/10.1038/s41594-021-00652-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00652-z
This article is cited by
-
In silico prediction of immune-escaping hot spots for future COVID-19 vaccine design
Scientific Reports (2023)
-
Structural dynamics in the evolution of SARS-CoV-2 spike glycoprotein
Nature Communications (2023)
-
Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection
Journal of Biomedical Science (2022)
-
COVID-19 vaccine update: vaccine effectiveness, SARS-CoV-2 variants, boosters, adverse effects, and immune correlates of protection
Journal of Biomedical Science (2022)
-
In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM
Nature Communications (2022)