Abstract
The current severe situation of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has not been reversed and posed great threats to global health. Therefore, there is an urgent need to find out effective antiviral drugs. The 3-chymotrypsin-like protease (3CLpro) in SARS-CoV-2 serve as a promising anti-virus target due to its essential role in the regulation of virus reproduction. Here, we report an improved integrated approach to identify effective 3CLpro inhibitors from effective Chinese herbal formulas. With this approach, we identified the 5 natural products (NPs) including narcissoside, kaempferol-3-O-gentiobioside, rutin, vicenin-2 and isoschaftoside as potential anti-SARS-CoV-2 candidates. Subsequent molecular dynamics simulation additionally revealed that these molecules can be tightly bound to 3CLpro and confirmed effectiveness against COVID-19. Moreover, kaempferol-3-o-gentiobioside, vicenin-2 and isoschaftoside were first reported to have SARS-CoV-2 3CLpro inhibitory activity. In summary, this optimized integrated strategy for drug screening can be utilized in the discovery of antiviral drugs to achieve rapid acquisition of drugs with specific effects on antiviral targets.
Similar content being viewed by others
Introduction
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which can lead to various symptoms including fever, cough, fatigue, shortness of breath, and loss of smell and taste1. Up to September 10, 2021, more than 223.2 million persons have been infected into COVID-19 across 192 countries or regions which resulted into 4,605,789 deaths2. Thus it will be urgent to seek for the effective treatment against COVID-19.
Since the outbreak of SARS-CoV-2, various antiviral compounds have been developed to treat COVID-193. At present, mainstream antivirus research has been conducted on the mechanism of virus replication, and many effective compounds have been discovered. A series of antivirals being tested against SARS-CoV-2 such as PF-073048144, remdesivir5, GC3766, apilimod7, nelfinavir8 and quinacrine9, displayed high antiviral activity in vitro. Some of them, such as remdesivir10, favipiravir11 and PF-07304814 are also under clinical investigation. However, no drugs have shown outstanding therapeutic effects in clinical trials. As the epidemic situation remains grim, there is still an urgent need for effective methods to discover valid antiviral drugs.
In addition to the mentioned chemical compounds, another important treatment approach is antiviral natural products (NPs) and herbal medicines. These herbal medicines as adjunctive treatment have been used to administrate the mild and moderate patients with coronavirus infection, including those caused by Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV and SARS-CoV-212,13,14. Traditional Chinese medicine (TCM), as a material basis for the application of NPs and herbal medicines under the guidance of theory, have been found to be an effective treatment for COVID-19. Previously, the "three-medicines and three-prescriptions (TMTP)" strategy was recommended as a prescribed formula by the State Council of China because TMTP has exhibited the remarkable therapeutic effects and no side effects, especially against COVID-1915. TMTP mainly include Jinhua Qinggan granules, Lianhua Qingwen granules and capsules, Xuebijing injection, Qingfei Paidu decoction, Huashi Baidu decoction and Xuanfei Baidu decoction, which have presented good clinical efficacy in the treatment of COVID-1916. This strategy halts the progression of the disease and actively alleviates and improves symptoms during the early and middle stages16,17.
As a combination of several compound medicines, TMTP contains abundant molecules that make the antiviral mechanism unclear. It is necessary to apply an efficient method that quickly and effectively finds the basis of antiviral substances from this valuable natural molecular library. In recent years, the application of in silico technology in drug discovery has achieved prominent success18,19, supplying sophisticated tools for screening promising lead compounds, predicting potential protein catalytic sites or revealing the mode of protein–ligand interactions. During this pneumonia epidemic, studies have utilized the favored approaches that target SARS-CoV-2 with high-throughput screening of large-scale molecular databases and obtaining potential antiviral drugs20,21. With the advancement of computer technology, the combination of computer-aided drug design (CADD) and artificial intelligence (AI) research has become a valuable tool to accelerate the slow process of drug discovery and restraint the expansion of R&D costs, expand the applicable system and improve the level of automation, followed by the development of CADD-based multithreaded in silico screening technology22,23,24,25. Within the framework of above idea, we proposed a multimodule integrated approach aimed at improving the lead compound screening accuracy and greatly reducing the time cost by fully maximizing the advantages of each module to achieve a semiautomatic pipeline.
Based on the above purposes, an integration of CADD, machine learning (ML) and similarity-based clustering methodologies and experimental validation was adopted in our study as an accurate and efficient means to filter potential anticoronavirus candidates from TMTP database against 3CLpro, the key target of viral replication26,27,28,29. We expeditiously discovered 5 NPs inhibitors, which further supports that the integrated strategy can accurately and quickly achieve the purpose of obtaining promising lead compounds, suggesting its practicality and worthiness for further optimizing the application form and systems.
Results
Workflow construction and molecular data base constitutions
This integrated screening workflow is divided into four parts (Fig. 1). First, a molecular library was constructed, and the affinity ranking was obtained through molecular docking. Then, cluster analysis was performed to reveal the molecular characteristics of high-affinity clusters and extract the top clusters. Furthermore, combined with high-throughput methods applied to determine the binding affinity and predict the level of activity, the molecules with both excellent properties were finally verified experimentally to determine effective compounds.
In the foremost step, we retrieved 49 Chinese medicinal materials involved in the 6 compound herbal formula in TMTP (Supplementary Table 1). First, excluding gypsum, which is mainly inorganic salt, 5464 SDF files of each chemical ingredient related to 46 Chinese medicinal materials were obtained through TCMSP database, the molecules of Herba Rhodiolae30 and Rhizoma Areactylodis Lanceae31 were excavated from literature, then 8 and 13 compounds were added from PubChem respectively. Second, we removing duplicates from 5485 compounds. Finally, 3272 compounds were obtained, which is the TMTP molecular database. This library include the Chinese herbal compound prescriptions and the representative Chinese medicines from TMTP as well as the main chemical compositions. On the other hand, we collected 301 of SARS-CoV and 84 of SARS-CoV-2 3CLpro inhibitors, which was performed as comprehensively as possible. The former was used to build ML models, and the latter were treated as a test set. A complete list of the molecules and related information for 3272 TMTP compounds library, compound libraries and SARS-CoV and SARS-CoV-2 3CLpro inhibitors is given in the Supplementary Data 1.
20 clusters divided from the TMTP compound library by cluster analysis
To classify the structural similarity of high-affinity molecules to further narrow the range of lead compounds, a total of 8 combinations between similarities of fingerprint maps and different cluster agglomeration methods were individually used for cluster analysis. Based on the agglomerative coefficient from agnes, we found that the combination of Euclidean and Ward2 exhibited the highest value (agglomerative coefficient = 0.975) compared with that of the other groups, and the agglomerative coefficients of the 8 groups are listed in Supplementary Table 2. Thus, we adopted the Euclidean and Ward2 combination to plot a clustering that contained 20 clusters (k = 20) (Supplementary Fig. 1, Supplementary Table 3).
Dominant clusters determined by means of molecular docking and ranking
Our molecular docking approach was used to obtain the binding ability of the TMTP molecular library (3272 molecules) with SARS-CoV-2 3CLpro, as well as the affinity score between positive inhibitors with SARS-CoV (301 of SARS-CoV 3CLpro inhibitors) or SARS-CoV-2 (301 of SARS-CoV and 84 of SARS-CoV-2 3CLpro inhibitors) 3CLpro for ML modeling. Docking analysis was carried out independently using the programs Autodock Vina, Glide, and MOE. Then, the exponential consensus ranking (ECR) strategy was implemented to reduce the number of false positives. This approach transformed docking scores of a single compound into a decimal number to indicate the comprehensive binding level for the target-ligand complex. Subsequent analyses were performed using ranking values instead of docking scores (Supplementary Data 1).
To compare the binding capacity to 3CLpro among clusters, we calculated and ranked the median, mean and quantile value, etc. of the molecular ranking in each cluster. Then, the dominant clusters were defined as those with a mean ranking value greater than 0.6 (Supplementary Table 4), and 9 dominant clusters were ultimately acquired. Among the 9 dominant clusters, the average ranking value of the royalblue cluster and brown cluster was greater than 0.7, indicating that these two clusters have higher target affinity.
Combining binding affinity of SPR and inhibitory activity prediction by ML analysis to narrow the range of hit compounds
In current study, surface plasmon resonance (SPR) was used to rapidly identify molecules in the dominant clusters that have the ability to bind to SARS-CoV-2 3CLpro. As a result, 21 molecules demonstrated high affinity for 3CLpro (Supplementary Table 5). ML was applied in parallel with SPR analysis to predict the 3CLpro inhibitory efficiency of compounds in the dominant clusters and further eliminate the molecules that would be nonspecifically bound in the SPR analysis. As previously described, the collected information on 3CLpro inhibitors (301 SARS-CoV and 84 SARS-CoV-2 inhibitors), including IC50, pIC50, SMILES, and CID, is shown in Supplementary Data 2. After the molecular docking process, we acquired the docking scores and molecular ranking values between the SARS-CoV 3CLpro inhibitors and 3CLpro of SARS-CoV and SARS-CoV-2 using three different software programs. Overall, no significant difference between docking scores or molecular ranking values of SARS-CoV and SARS-CoV-2 was observed, which was ascribed to the high homology of the SARS-CoV and SARS-CoV-2 3CL proteins32. We calculated the similarity index (0.710) of two proteins binding or activity based on the docking matrix. Then, the predicted IC50 of SARS-CoV-2 3CLpro was computed based on the Eq. (2). By means of Rcpi, the molecular descriptors of 301 compounds were extracted as the quantitative structure (Supplementary Data 2). Thereafter, we constructed the quantitative relationship between structure and activity by random forest (RF) and support vector machine (SVM) training classification models. The activity of the 84 compounds for SARS-CoV-2 3CLpro was tested using the training model. To deal with the imbalance training datasets in the RF and SVM algorithm, we used the method of additional sampling that was conducted after resampling (usually to resolve class imbalances). The results of comparison among four resampling methods in these two algorithms had shown the area under curve (AUC) value of smote method of RF was higher than the other methods (RF: AUCsmote = 0.87, AUCdown = 0.83, AUCregional = 0.85, AUCweight = 0.86; SVM: AUCsmote = 0.81, AUCdown = 0.79, AUCregional = 0.81, AUCweight = 0.80). Thus, we incorporated the smote algorithm and cross-validation methods into the model in the train function. To further compare the RF and SVM analysis, the training model was tested with data of SARS-CoV-2 (81) and multiple evaluation metrics including the AUC, recall and precision value were calculated. We found that the AUC of receiver operating characteristic curve (ROC) in RF was higher than that of SVM (RF: AUC = 0.69, Precision = 0.24, Recall = 0.5; SVM: AUC = 0.59, Precision = 0.27, Recall = 0.30). Suggesting that the predicted inhibitory value calculated by the RF method was closer to the experimental value than that calculated by the SVM method.
Finally, we predicted the activity of 9 dominant clusters; here, a predicted value represented as possibility of positive result which have been computed with predict function and the selected type as prob, that greater than 0.5 was considered to have an inhibitory effect, and vice versa. A total of 156 compounds from 816 compounds in the dominant cluster were predicted to be active via ML analysis. A complete list of the predicted values can be found in Supplementary Data 3.
We combined the binding ability results from SPR analysis and the predicted activity results from ML analysis. The 11 NPs that have shown positive both SPR and ML analysis were considered potential inhibitor candidates and utilized for further experiments. Interestingly, these high-activity compounds were enriched in the brown, midnightblue and red clusters (Supplementary Table 6). In the above clusters, the brown cluster mainly contains flavonoids and their glycosides. The midnightblue cluster is composed of dammarane and oleanane or their derivative parent nucleus and corresponding glycosides. The compounds in the red cluster are composed of polyhydroxy conjugated systems such as hydroxytyrosol and caffeic acid to connected with sugar units. These types of compounds often exhibit a wide range of biological activities and have also been used in the field of anti-virus33,34.
5 NPs identifying as potent inhibitors of SARS-CoV-2 3CLpro in vitro
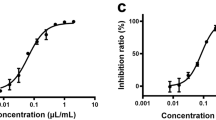
Eleven compounds selected by the virtual screening and ML analysis were subsequently tested using the inhibition assay against SARS-Cov-2 3CLpro. After the initial screening, only five compounds at a concentration of 100 μM demonstrate over 50% inhibitory active against the enzyme. These compounds were able to achieve inhibition at lower concentrations. According to results shown in Fig. 2, narcissoside (MOL003686), kaempferol-3-O-gentiobioside (MOL012143), rutin (MOL000415), vicenin-2 (MOL001543) and isoschaftoside (MOL004958) presented IC50 values of 38.142, 35.892, 31.259, 38.856 and 30.220 μM, respectively. Remarkably, they are all flavonoids. The results of affinity screening by SPR showed that flavonoids accounted for 10 of the 21 compounds and their KD values ranged from 1.525 to 12.46, exhibited strong affinity with 3CLpro (Fig. 3). As demonstrated in Table 1, the KD values are well correlated with IC50 values. Notably, kaempferol-3-o-gentiobioside, vicenin-2 and isoschaftoside were reported as anticoronavirus candidates for the first time due to their inhibition of 3CLpro of SARS-CoV-2. They are very promising for further research to develop compounds with high inhibition efficiency.
Molecular dynamics simulation revealed the stable binding mode of the 5 selected drugs with SARS-CoV-2 3CLpro
The dynamic binding interactions of the five compounds with inhibitory activity were analyzed, and 100 ns molecular dynamics (MD) simulations of ligand–protein complexes were performed. The root mean square deviation (RMSD) of the ligand trajectory was analyzed, revealing that the complexes rapidly reached equilibrium within the first 5 ns of the simulation (Fig. 4a), with each value lying between 1.5 and 3.5 Å. Narcissoside and vicenin-2 fluctuated greatly, indicating a flexible bingding to the active site of 3CLpro. In contrast, compounds kaempferol-3-O-gentiobioside, rutin, and isoschaftoside are more fixed. On the other hand, the degree of configuration change of these compounds in the binding pocket is relatively stable, and the RMSD is less than 0.5 (Fig. 4d).
The RMSD values and contribution of various energy items to binding free energy in drug-3CLpro simulation. (a) Root mean square deviation (RMSD) of the 5 3CLpro-ligand complexes from the 100 ns MD simulations. Narcissoside (blue), kaempferol-3-O-gentiobioside (red), rutin (black), vicenin-2 (green) and isoschaftoside (Violet). (b) Residues with a high contribution to the total binding energy during the MD simulation of the 5 protein-inhibitor complexes. Narcissoside (blue), kaempferol-3-O-gentiobioside (red), rutin (black), vicenin-2 (green) and isoschaftoside (violet). (c) The general structure of narcissoside, kaempferol-3-O-gentiobioside and rutin. (d) Root mean square deviation (RMSD) of the 5 ligand from the 100 ns MD simulations. Narcissoside (blue), kaempferol-3-O-gentiobioside (red), rutin (black), vicenin-2 (green) and isoschaftoside (Violet). (e) The contribution of various energy items to the total binding energy of 3CLpro inhibitors. Narcissoside (blue), kaempferol-3-O-gentiobioside (red), rutin (black), vicenin-2 (green) and isoschaftoside (violet). f The general structure of vicenin-2 and isoschaftoside.
To explore the binding affinity of each ligand to 3CLpro, the binding free energy was calculated based on MM/PBSA (Table 2). Van der Waals (ΔEvdW) and electrostatic (ΔEele) interactions make major contributions to the binding free energy (Fig. 4e). We observed that rutin exhibited the highest binding affinity to 3CLpro, followed by kaempferol-3-O-gentiobioside, isoschaftoside, vicenin-2 and narcissoside. Analysis of the energy decomposition results of the five compounds suggested that the residues Thr25, Thr26, Ley27, His41, Cys44, Tgr45, Ser46, Met49, Asn142, Gly143, Cys145, His163, His164, Met165, Asp187 and Gln189 mainly contributed to hydrophobic and electrostatic interactions in the 3CLpro-ligand complex (Supplementary Table 7).
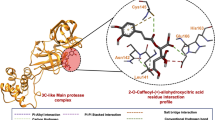
Specifically, from the analysis of the binding interactions, narcissoside showed the least electrostatic interaction (− 21.59 kcal/mol), forming hydrogen bonds with Ser46, Gly143, His164, and Glu166. Kaempferol-3-O-gentiobioside forms multiple hydrogen bonds with Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, Glu166, Pro168, and Arg188. Rutin forms hydrogen bond interactions with Thr26, Tyr54, Phe140, Asn142, Gly143, Glu166, and Gln189. Vicenin-2 demonstrated the highest number of H-bonds, forming hydrogen bonds with Thr26, Phe140, Asn142, Gly143 and Glu166. In the analysis of binding energy with isoschaftoside, the contribution of electrostatic interactions to the total binding energy was − 66.58 kJ/mol, which was highest among the 5 compounds, forming H-bond interactions with Thr26, Tyr54, Phe140, Asn142, Gly143 and Glu166, Gln189. The above analysis suggested that flavonoid glycosides provided higher flexibility after forming chains with sugars because of their rotatable bonds, which can bind into pockets and form abundant hydrogen bonds with some key residues. From the perspective of amino acid energy decomposition (Fig. 4b), the compound has a strong interaction with His41, Met49, and Cys145. His41 and Met49 are also the active site residues of 3CLpro35. To facilitate the analysis, we first colored the region of the residues His41 and Met49, and then divided the five flavonoids into two categories according to their structural similarity. The active cavity of 3CLpro presented strong hydrophobicity, while the aromatic ring of the flavonoid aglycone provided the main hydrophobic energy contribution in the site. For type A (Fig. 4c), narcissoside, as the only inhibitor with methoxy group. The group has the function of enhancing hydrophobic action of ligand (-58.32 kcal/mol), making the benzene ring easily inserting into the hydrophobic region of the cavity (Fig. 5a,f), resulting in the overall structure extending outside of the cavity and reducing the interaction with residues, eventually reducing the contribution of the binding free energy. In contrast, the flavonoid skeleton of kaempferol-3-O-gentiobioside is close to the cavity (Fig. 5b,g). Furthermore, rutin is inserted into the cavity (Fig. 5c,h), which makes the binding tighter and presents the lowest binding free energy (Table 2). For type B (Fig. 4f), the overall structure shifted in the active pocket due to prolongation of the rigid flavonoid part in vicenin-2 (Fig. 5d,i) and isoschaftoside (Fig. 5e,j), resulting in the distance from the active site being farther than that for kaempferol-3-O-gentiobioside and rutin. However, they did not demonstrate much difference in their total binding free energies.
Binding mode of 5 NPs inhibitors to SARS-CoV-2 3CLpro. The protein and ligand are shown in gray and green, respectively. H-bond interactions are demonstrated by yellow dotted lines and residues forming H-bonds are shown in cyan. The surface of the protein is represented in cyan. The residues His41 and Met49 are highlighted in red to show the binding posture of the inhibitor. (a) Binding interaction of narcissoside. (b) Binding interaction of kaempferol-3-O-gentiobioside. (c) Binding interaction of rutin. (d) Binding interaction of vicenin-2. (e) Binding interaction of isoschaftoside. (f) Binding posture of narcissoside. (g) Binding posture of kaempferol-3-O-gentiobioside. (h) Binding posture of rutin. (i) Binding posture of vicenin-2. (j) Binding posture of isoschaftoside.
Notably, from the analysis of the binding interaction, with the key residues, we observed that the interaction strength between His41 and Met49 with the ligand was positively correlated with the affinity of the ligand and 3CLpro binding. In addition, the total energy (Supplementary Table 8) of residues based on the region of His41 to Met49 also exhibited this rule. On the other hand, ΔGTot calculated by MM/PBSA also matched the rank of the molecule (Table 2). The clear binding pattern and significant inhibitory activity of these five flavonoids against 3CLpro indicated that they are promising candidates for anti-SARS-CoV-2 activity. These results prove the correctness of our screening strategy.
Discussion
To fight the epidemic and obtain effective antiviral drugs from a reservoir of herbal medicines, we designed an integrated pipeline workflow for NP screening. Our integrated strategy combining each submodule into the best workflow and fulfilling the optimization of function therefore exhibited a positive impact from rapid and accurate acquisition of lead compounds to subsequent structural optimization guidance. It is worth noting that the selection and optimization of submodules can be flexibly changed; this integrated strategy may not be limited to NPs screening. Furthermore, this strategy has the potential to derive automated pipelines from prototype workflows to improve the convenience of use while ensuring the accuracy of screening.
The screening pipeline was based on the binding affinity between molecules and targets. To avoid the affinity error caused by limitations of an algorithm of a single software which eliminate compounds that have true binding ability, we selected 3 commonly used docking software programs and converted docking scores into rankings of affinity trends by means of consensus analysis. Attributed to an "or" condition, the ECR assay can reduce the impact of extreme values in software scoring36, which achieved the fault tolerance of the discrepancy from different scoring functions, that it is very suitable for the research strategy of our study.
When confronted with a large compound library, effective cluster analysis can directly reflect the structural characteristics of molecules or clusters with high affinity and indicate the types of functional groups with high contribution to the corresponding complexes. This helps to guide the subsequent structural optimization and quickly eliminate the set of compounds with poor binding ability to the target. Consequently, similarity-based compound clustering is crucial in pipelines. Among the hierarchical and nonhierarchical clustering methods, Ward's and Jarvis-Patrick are known to be effective algorithms for chemical structure clustering37,38. As expected, in the course of practice, Jarvis-Patrick produced too many singletons and a small number of large clusters39, which is completely inconsistent with our requirements. For the similarity calculation, Tanimoto coefficients and Euclidean distance are the most widely used to evaluate how similar two molecules are to each other40,41. Then, we combined them with 4 commonly used hierarchical clustering algorithms and compare the calculated corresponding 8 sets of agglomerative coefficient. Thereafter, we found that Euclidean and Ward2 is the best matches to obtain a cluster of uniform internal structure characteristics, and the number of clusters can also be optimized for specific systems. In summary, this high-precision clustering is suited for but not limited to natural products, and it is worthy of promotion.
As an effective tool for predicting the structure–activity relationship, ML has been widely used in threaded approach42,43. The present ML-based activity prediction was capable of describing active molecules from the TMTP database even with a small-volume training set (301 compounds). We calculated the predicted value of SARS-CoV-2 3CLpro from the collected SARS-CoV 3CLpro inhibitory activity according to the similarity matrix between docking data of two target proteins. To build an effective model, we increased molecular character number of the training compound as an independent variable and covered the docking data. Moreover, we compared the predictive accuracy of the SVM and RF methods integrated into the cross validation analysis and found that the AUC value of random forest was higher than SVM, which provided the evidence for the candidates drug discovery by RF-based activity prediction.
Current cluster analysis accurately placed flavonoids into a subset and further obtained the 5 compounds with inhibitory activity of 3CLpro. As compounds characterized by 2-phenyl-benzyl-γ-pyrone nucleus, flavonoids are particularly valuable NPs that possess anti-inflammatory44, antioxidant45, anti-microbial46, and even antiviral activities. In a study of inhibitors against coronavirus, rhoifolin, pectolinarin, herbacetin47 and amentoflavone48 were demonstrated to block the function of SARS-CoV 3CLpro. The glycoside derivatives of kaempferol also proved to be virus release inhibit agents by blocking the 3a channel49. During the COVID-19 outbreak, narcissoside50 and rutin29 also exhibited the inhibitory effects on main protease of SARS-CoV-2, which is confirmed the practicality of our screening strategy.
The limitations of this study have been presented. First, on account of the few reports of tested compounds, a relatively small sample training set in the ML analysis was available. When more positive drugs are involved, the accuracy of ML activity prediction can be further improved. Second, the 5 NPs have not been evaluated the antiviral activity against SARS-CoV-2 by in vivo experimental and preclinical data due to the powerful invasion ability of SARS-CoV-2.
In conclusion, this study successfully employed an integrated screening strategy to identify 5 potential inhibitors of SARS-CoV-2 3CLpro from a NPs library composed of clinically effective herbal medicines. On the basis of this research, further research is worth pursuing to produce derivatives that can produce better inhibitors. The high efficiency and accurate characteristics of this strategy greatly shorten the hit cycle of lead compounds in the process of drug discovery for acute diseases and accelerate the process of drug development. We recommend that this integrated screening strategy be applied to other targets that urgently need effective drugs.
Methods
Construction of TMTP chemical constituent databases
Except for gypsum whose main component is inorganic salt, the SDF files of each chemical ingredient related to 46 Chinese medicinal materials were obtained through TCMSP database (Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform, http://lsp.nwu.edu.cn/tcmsp.php). By consulting the related literature of Herba Rhodiolae and Rhizoma Areactylodis Lanceae, SDF files were downloaded through PubChem respectively. The TMTP molecular database was obtained after the database deduplication. Then, the OpenBabel toolkit was used to convert the mol2 file of each molecule into unified SDF, pdbqt and SMILES file formats to prepare for molecular docking. Subsequently, we retrieved the absorption, distribution, metabolism and excretion (ADME) properties data from TCMSP database, containing molecular weight (MW), oral bioavailability (OB), number of hydrogen-bond acceptors (HBA), number of hydrogen-bond donors (HBD), etc. Other molecular descriptors, including atom additive logP (ALogP), atom molar refractivity values (AMR), and topological polar surface area (TPSA), were calculated based on the Rcpi in the R platform. All the discriptors were listed in Excle table. After deduplicating the SMILES file, the retained molecular discriptors entries and SDF files constituted a compound library for subsequent analysis.
Molecular docking
To accurately predict docking poses, three different molecular docking programs, AutoDock Vina51 (version 1.1.2), Maestro (version 11.4, Schrödinger, LLC, New York, NY, 2021), and molecular operating environment software (MOE, Chemical Computing Group, version 2019.0101), were used to detect the binding capability between diverse compounds and SARS-CoV or SARS-CoV-2 3CLpro. The protein and ligand were prepared for the docking process. First, the crystal structural file of SARS-CoV and SARS-CoV-2 3CLpro were downloaded from protein database (PDB ID 3V3M52, 6LU753). For the target protein, the preparation included carrying out the assign bond orders, hydrogenation, treatment of disulfide bonds, metal ions, and removal of water molecules, heteroatoms with default settings in three software. For ligand preparation, apart from AutoDock Vina which minimized the compound’s energy in Chem3D software, Maestro and MOE were carried out with inner LigPrep and Energy Minimize protocol respectively, to generated the correct form, and all the hydrogen atoms and the torsion information were added. To ensure the uniformity of different software at the docking position, the binding site box of 3CLpro coincided with binding site of the original inhibitors. Docking analysis was conducted with default protocol in AutoDock Vina, the Extra precision (XP) was used in GlideScore scoring functions54, and the Induced Fit module was chosen in MOE. The PyMOL Molecular Graphics System (version 2.0, Schrödinger, LLC) was used to visualize the docking posture of compounds at the binding pocket of 3CLpro.
Consensus analysis
To combine results from several docking programs, we adopted ECR strategy proposed by Karen Palacio-Rodríguez et al.36 (2019) to assigned a rank to each molecule based on the docking scores of the molecules provided by different docking program. As shown in Eq. (1), \({\upsigma }\) represented the expected value of an exponential distribution and was assigned to be 10. \(s_{n}\) referd to the compound docking score given by each software. The ultimate rank value of each molecule was defined as the sum of all the exponential scores \(p\left( {s_{n} } \right)\), which was a positive correlation between the rank value and affinity of compound against target protein.
Cluster analysis
To identify homogeneous and distinct groups, or similar objects in TMTP NPs datasets, we performed clustering analysis with the ChemmineR and WGCNA packages. First, we collected the SDF file of each molecule produced by the OpenBabel software. By means of readMolFromSDF, we converted the SDF files into mol files. The SDF files were loaded into the ChemmineR for calculating atom pair fingerprints (APfp) of all compounds, which were used for calculate the structural similarity between the different compounds55,56. We adopted the Tanimoto coefficients and Euclidean distance for computing distance or dissimilarity metrics based on the fingerprint of TMTP natural products. In this proccess, we mainly utilized the sdf2ap and fpSim function from ChemmineR package. Firstly, we extracted atom pair fingerprints from 3272 NPs sdf files through sdf2ap function. Then, we calculated pairwise compound structure comparisons from fingerprint dataset using fpSim function. The fingerprint-based Tanimoto or Euclidean similarity matrix were computed and the other parameters were set as default values. To classify the different compounds into a series of the relative number of clusters, we concentrated on single, complete, average, and Ward's algorithms to map the strength of the clustering results. Additionally, the dendrograms of 8 hclust approaches were plotted, and the agnes from dendextend computing the agglomerative coefficient was used to measure the amount of clustering structure found (values closer to 1 suggest a strong clustering structure). To clearly delineate the different clusters, we integrated the WGCNA, which can effectively assign different modules and are represented as visualized colors. The functions of cutreeDynamic and labels2colors were further used to investigate the best clustering results.
Surface plasmon resonance analysis
The binding studies were performed at 25 °C on a Biacore T200 instrument at a flow rate of 30 μL/min in running buffer composed of PBS (pH 7.4) and 3 mM EDTA. CM7 chips activated in a 10 min injection procedure with a mixture of EDC (1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide)/NHS (Nhydroxysuccinimide) (0.2 M/0.05 M) and immobilized with anti-histone antibody until levels of immobilization were between 15,000 and 20,000 RU. The chip was then deactivated with a 7 min injection of 1 M ethanolamine (pH 8.0). The 3CLpro-his protein was then applied on the chip to reach typically levels between 4000 and 5000 RU. The binding activity and stability of proteins to ligands were tested at the end of each compound injection. The association and dissociation phases of tested ligands were monitored for 120 s each. The final binding experiment with small molecule ligands was performed in the above running buffer supplemented with 5% DMSO. PBS, EDTA, CM7 chip, EDC, NHS and ethanolamine were obtained from GE, 3CLpro-his was obtained from Kangma- Healthcodea.
Machine learning analysis
3CLpro Positive inhibitors of SARS-CoV and SARS-CoV-2 for individual modeling and validation via ML were acquired from the literature, related IC50 were collected, and the SDF files was downloaded from Pubchem or generated by ChemDraw, as well as SMILES files. Then, each 3CLpro inhibitor of SARS-CoV was docked with the 3CLpro of SARS-CoV and SARS-CoV-2 separately, as described in the docking procedure above, rank of a molecule was constructed from the docking score matrix. To eliminate the inconvenience of calculation and data comparison caused by the orders of magnitude difference between the data, pIC50 was applied in calculation instead of IC50. After that, the inhibitory efficiency in the data set was converted to two classes: active (pIC50 ≥ 6) and inactive (pIC50 < 6). Then, the similarity index (SI) between docking score of compound and 3CLpro complex in SARS-CoV and SARS-CoV-2 was calculated via SMI from MatrixCorrelation57. To build the predicted model, we proposed the Eq. (2) to transform the SARS-CoV IC50 to predicted IC50 of the SARS-CoV-2 inhibition efficiency. Subsequently, a classification model based on the quantitative structure and activity relationship of SARS-CoV-2 was constructed.
The inhibitory compounds targeting SARS-CoV-2 3CLpro and the related IC50 were validated as a test data set. By means of Rcpi, we extracted the molecular descriptors of the validated compounds, including ALogP, square of AlogP (ALogp2), AMR, atomic polarizabilities (apol), eccentric connectivity index (ECCEN), topological polar surface area (TopoPSA), MW, weiner path number (WPATH), weiner polarity number (WPOL) and the sum of the squares of atom degree over all heavy atoms (ZagrebIndex). The molecular descriptors and docking value of the training data sets were input as independent variables, and the predicted IC50 values were input as dependent variables. We selected the random forest (RF) and support vector machine (SVM), which are widely used ML methods58,59. We established the machine learning analysis by the caret package. Repeated cross validation (times = 5, fold = 10) were used as the cross-validation strategies. And we optimized the important parameters including sigma, C and weight which were used to select the optimal model. The main hyperparameters of SVM contain sigma (sigma = 0.1), cost (C = 1) and weight (weight = 3) while the hyperparameters of RF include mtry (mtry = 2). Meanwhile, considering that the imbalanced training dataset, we explored different algorithms, including original, weighted, down and smote algorithms, in the training stage. The accuracy of two classification model was evaluated by the AUC value of ROC. The recall and precision values were calculated with confusionMatrix function. Based on the AUC value, we selected the optimal methods to predict the inhibition efficiency of natural products from dominant clusters.
SARS-CoV-2 3CLpro inhibition assay
The inhibition assay of SARS-CoV-2 3CLpro was carried out based on the reported method60. In the initial step, 0.5 µg of SARS-CoV-2 3CLpro was preincubated with 100 µL of 200 µM test compound at room temperature for 15 min. Then, the reaction was triggered after the addition of 10 µM Dabcyl-KTSAVLQSGFRKME-Edans (GL Biochem). The fluctuation of fluorescence intensity was monitored on a GENios microplate reader (Tecan), where the excitation wavelength was 340 nm and the emission wavelength was set to 490 nm. Control reactions were performed under the same condition, but the compounds or enzymes were excluded from the reaction system. The median inhibitory concentration (IC50) values against SARS-CoV-2 3CLpro was calculated by nonlinear regression analysis via GraphPad Prism 7.03 (GraphPad Software, San Diego, CA, USA).
Molecular dynamics simulation and binding free energy calculation
MD simulations of the screened natural inhibitors of 3CLpro were performed with Amber1461 to evaluate their binding interaction patterns with 3CLpro. The protein–ligand complexes were used as the initial structure for subsequent MD simulations. The ligand and protein were treated with General Amber Force Field (GAFF62) and FF14SB63 respectively. For the amino acid residues of the protein, the default protonation state in Amber14 was adopted, and the hydroprocessing was carried out using tleap module. By means of the Gaussian09 software package64, the Lee–Yang–Parr correlation functional (B3LYP)/6-31G** was carried out to optimized all inhibitors. The restrained electrostatic potential (RESP) charges as partial charges for molecules were calculated by fitting with the standard RESP procedure implemented in the Antechamber module of the Amber14. And molecular dynamics simulations for all complexes employ the PMEMD program in Amber14. After adding counter ions to each complex to maintain the neutrality of the system, the entire system was contained to a TIP3P rectangular water box. Furthermore, energy minimization was performed by steepest descent method of 2500 steps and conjugate gradient method of 2500 steps. Subsequently, same methods were used to optimize the unconstrained system. The Particle mesh Ewald (PME) was performed in the MD simulation to deal with the long-range electrostatic interaction, and the SHAKE algorithm was used to constrain all the bonds connected to the hydrogen atom and the time step was set to 2 fs. Then set a cutoff value of 10 Å for non-bonding interactions. The constrained whole system was heated from 0 to 300 K in 60 ps at a constant volume, subsequently, the solvent density was balanced in a constant pressure and thermostatic system (T = 300 K, P = 1 ATM). Followed by 100 ns of MD simulations control at constant pressure and frames were saved at 1 ps intervals (50,000 frames totally) for subsequent MM/PBSA analysis at last. In order to obtain the RMSD, the trajectory was generated from MD simulation via cpptraj module then analyzed with the VMD (1.9.3) program65. OriginPro (Version 2021 Learning Edition. OriginLab Corporation, Northampton, MA, USA.) was used for plot.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files.
Code availability
The authors declare that all code supporting the findings of this study are available from the corresponding author on reasonable request.
References
Grant, M. C. et al. The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): A systematic review and meta-analysis of 148 studies from 9 countries. PLoS ONE 15, e0234765 (2020).
Dong, E., Du, H. & Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet. Infect. Dis. 20, 533–534 (2020).
Rosa, S. G. V. & Santos, W. C. Clinical trials on drug repositioning for COVID-19 treatment. Rev. Panam. Salud. Publica. 44, e40 (2020).
de Vries, M. et al. Comparative study of a 3CL pro inhibitor and remdesivir against both major SARS-CoV-2 clades in human airway models. bioRxiv. Preprint at http://biorxiv.org/content/early/2020/08/28/2020.08.28.272880.abstract (2020).
Gordon, C. J. et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 295, 6785–6797 (2020).
Vuong, W. et al. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 11, 4282 (2020).
Riva, L. et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 586, 113–119 (2020).
Yamamoto, N., Matsuyama, S., Hoshino, T. & Yamamoto, N. Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro. bioRxiv. Preprint at https://www.biorxiv.org/content/biorxiv/early/2020/04/08/2020.04.06.026476.full.pdf (2020).
Salas Rojas, M. et al. Quinacrine, an antimalarial drug with strong activity inhibiting SARS-CoV-2 viral replication in vitro. Viruses 13, 121 (2021).
Beigel, J. H. et al. Remdesivir for the treatment of covid-19 - final report. N. Engl. J. Med. 383, 1813–1826 (2020).
Joshi, S. et al. Role of favipiravir in the treatment of COVID-19. Int. J. Infect. Dis. 102, 501–508 (2021).
Lin, S. C. et al. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect. Dis. 17, 144 (2017).
Leung, P. C. The efficacy of Chinese medicine for SARS: A review of Chinese publications after the crisis. Am. J. Chin. Med. 35, 575–581 (2007).
Shahrajabian, M. H., Sun, W., Shen, H. & Cheng, Q. Chinese herbal medicine for SARS and SARS-CoV-2 treatment and prevention, encouraging using herbal medicine for COVID-19 outbreak. Acta Agric. Scand. Sect. B 70, 437–443 (2020).
The state council information office, Press Conference of the Joint Prevention and Control Mechanism http://www.gov.cn/xinwen/gwylflkjz92/index.htm (2020).
Huang, Y. F., Bai, C., He, F., Xie, Y. & Zhou, H. Review on the potential action mechanisms of Chinese medicines in treating Coronavirus Disease 2019 (COVID-19). Pharmacol. Res. 158, 104939 (2020).
Hu, K. et al. Efficacy and safety of Lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus disease 2019: A multicenter, prospective, randomized controlled trial. Phytomedicine https://doi.org/10.1016/j.phymed.2020.153242 (2020).
Reynolds, C. H. Impact of computational structure-based methods on drug discovery. Curr. Pharm. Des. 20, 3380–3386 (2014).
Nicolaou, C. A. et al. Idea2Data: Toward a new paradigm for drug discovery. ACS Med. Chem. Lett. 10, 278–286 (2019).
Fischer, A., Sellner, M., Neranjan, S., Smieško, M. & Lill, M. A. Potential inhibitors for novel coronavirus protease identified by virtual screening of 606 million compounds. Int. J. Mol. Sci. 21, 3626 (2020).
Elmezayen, A. D., Al-Obaidi, A., Şahin, A. T. & Yelekçi, K. Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. https://doi.org/10.1080/07391102.2020.1758791 (2020).
Gupta, V. et al. Multi-dimensional computational pipeline for large-scale deep screening of compound effect assessment: An in silico case study on ageing-related compounds. npj Syst. Biol. Appl. 5, 42 (2019).
Samdani, A. & Vetrivel, U. POAP: A GNU parallel based multithreaded pipeline of open babel and AutoDock suite for boosted high throughput virtual screening. Comput. Biol. Chem. 74, 39–48 (2018).
Harnie, D. et al. Scaling machine learning for target prediction in drug discovery using Apache Spark. Future Gener. Comput. Syst. 67, 409–417 (2017).
Krivák, R. & Hoksza, D. P2Rank: Machine learning based tool for rapid and accurate prediction of ligand binding sites from protein structure. J. Cheminf. 10, 39 (2018).
Ziebuhr, J., Snijder, E. J. & Gorbalenya, A. E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81, 853–879 (2000).
Kumar, V., Tan, K. P., Wang, Y. M., Lin, S. W. & Liang, P. H. Identification, synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 24, 3035–3042 (2016).
Wang, L. et al. Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical synthesis, biological evaluation, molecular docking and 3D-QSAR study. Eur. J. Med. Chem. 137, 450–461 (2017).
Jo, S., Kim, S., Kim, D. Y., Kim, M. S. & Shin, D. H. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzyme. Inhib. Med. Chem. 35, 1539–1544 (2020).
Ma, Y.-C. et al. Simultaneous quantification of polyherbal formulations containing Rhodiola rosea L. and Eleutherococcus senticosus Maxim. using rapid resolution liquid chromatography (RRLC). J. Pharm. Biomed. Anal. 55, 908–915 (2011).
Zhang, W.-J. et al. Atractylodis Rhizoma: A review of its traditional uses, phytochemistry, pharmacology, toxicology and quality control. J. Ethnopharmacol. 266, 113415 (2021).
Tahir Ul Qamar, M., Alqahtani, S. M., Alamri, M. A. & Chen, L. L. Structural basis of SARS-CoV-2 3CL(pro) and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 10, 313–319 (2020).
Song, X., Yuan, G., Li, P. & Cao, S. Guanidine-containing polyhydroxyl macrolides: Chemistry, biology, and structure-activity relationship. Molecules 24, 3913 (2019).
Padhye, S. et al. Perspectives on chemopreventive and therapeutic potential of curcumin analogs in medicinal chemistry. Mini-Rev. Med. Chem. 10, 372–387 (2010).
Gul, S. et al. In silico identification of widely used and well-tolerated drugs as potential SARS-CoV-2 3C-like protease and viral RNA-dependent RNA polymerase inhibitors for direct use in clinical trials. J. Biomol. Struct. Dyn. https://doi.org/10.1080/07391102.2020.1802346 (2020).
Palacio-Rodríguez, K., Lans, I., Cavasotto, C. N. & Cossio, P. Exponential consensus ranking improves the outcome in docking and receptor ensemble docking. Sci. Rep. 9, 5142 (2019).
Varin, T., Bureau, R., Mueller, C. & Willett, P. Clustering files of chemical structures using the Szekely-Rizzo generalization of Ward’s method. J. Mol. Graphics Modell. 28, 187–195 (2009).
Malhat, M. G., Mousa, H. M. & El-Sisi, A. B. in 2014 9th International Conference on Informatics and Systems DEKM-61–66 (2014).
Khanna, V. & Ranganathan, S. Molecular similarity and diversity approaches in chemoinformatics. Drug Dev. Res. 72, 74–84 (2010).
Raymond, J. W., Blankley, C. J. & Willett, P. Comparison of chemical clustering methods using graph- and fingerprint-based similarity measures. J. Mol. Graph. Model. 21, 421–433 (2003).
Downs, G. M., Willett, P. & Fisanick, W. Similarity searching and clustering of chemical-structure databases using molecular property data. J. Chem. Inform. Comput. Sci. 34, 1094–1102 (1994).
Dey, S., Luo, H., Fokoue, A., Hu, J. & Zhang, P. Predicting adverse drug reactions through interpretable deep learning framework. BMC Bioinf. 19, 476 (2018).
Mayr, A., Klambauer, G., Unterthiner, T. & Hochreiter, S. DeepTox: Toxicity prediction using deep learning. Front. Environ. Sci. 3, 80 (2016).
Serafini, M., Peluso, I. & Raguzzini, A. Flavonoids as anti-inflammatory agents. Proc. Nutr. Soc. 69, 273–278 (2010).
Pietta, P.-G. Flavonoids as antioxidants. J. Nat. Prod. 63, 1035–1042 (2000).
Xie, Y., Yang, W., Tang, F., Chen, X. & Ren, L. Antibacterial activities of flavonoids: Structure-activity relationship and mechanism. Curr. Med. Chem. 22, 132–149 (2014).
Jo, S., Kim, S., Shin, D. H. & Kim, M. S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme. Inhib. Med. Chem. 35, 145–151 (2020).
Ryu, Y. B. et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg. Med. Chem. 18, 7940–7947 (2010).
Schwarz, S. et al. Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Med. 80, 177–182 (2014).
Dubey, K. & Dubey, R. Computation screening of narcissoside a glycosyloxyflavone for potential novel coronavirus 2019 (COVID-19) inhibitor. Biomed. J. 43, 363–367 (2020).
Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Jacobs, J. et al. Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J. Med. Chem. 56, 534–546 (2013).
Jin, Z. et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293 (2020).
Friesner, R. A. et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J. Med. Chem. 49, 6177–6196 (2006).
Bajusz, D., Racz, A. & Heberger, K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations?. J. Cheminf. 7, 20 (2015).
O’Boyle, N. M. & Sayle, R. A. Comparing structural fingerprints using a literature-based similarity benchmark. J. Cheminf. 8, 36 (2016).
Indahl, U. G., Næs, T. & Liland, K. H. A similarity index for comparing coupled matrices. J. Chemom. 32, e3049 (2018).
Manavalan, B., Shin, T. H. & Lee, G. DHSpred: Support-vector-machine-based human DNase I hypersensitive sites prediction using the optimal features selected by random forest. Oncotarget 9, 1944–1956 (2018).
Inglada, J., Vincent, A., Arias, M. & Marais-Sicre, C. Improved early crop type identification by joint use of high temporal resolution SAR and optical image time series. Remote Sens. 8, 362 (2016).
Chen, L. et al. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 79, 7095–7103 (2005).
Case, D. A. et al. AMBER 14 (University of California, 2014).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
Maier, J. A. et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Gaussian 09, Revision D.01, Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G. A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H. P., Izmaylov, A. F., Bloino, J., Zheng, G., Sonnenberg, J. L., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Montgomery, Jr. J. A., Peralta, J. E., Ogliaro, F., Bearpark, M., Heyd, J. J., Brothers, E., Kudin, K. N., Staroverov, V. N., Keith, T., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A., Burant, J. C., Iyengar, S. S., Tomasi, J., Cossi, M., Rega, N., Millam, J. M. Klene, M., Knox, J. E., Cross, J. B., Bakken, V., Adamo, C., Jaramillo, J., Gomperts, R., Stratmann, R. E., Yazyev, O., Austin, A. J., Cammi, R., Pomelli, C., Ochterski, J. W., Martin, R. L., Morokuma, K., Zakrzewski, V. G., Voth, G. A., Salvador, P., Dannenberg, J. J., Dapprich, S., Daniels, A. D., Farkas, O., Foresman, J. B., Ortiz, J. V., Cioslowski, J. & Fox, D. J. Gaussian, Inc., Wallingford CT (2013).
Humphrey, W., Dalke, A. & Schulten, K. V. M. D. Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Acknowledgements
We acknowledge the financial support from the National Natural Science Foundation of China (31871281), Shanghai Municipal Health Commission (2020XGKY12) and Scientific Research Foundation for Advanced Talents of Shanghai University of Traditional Chinese Medicine.
Author information
Authors and Affiliations
Contributions
Q.W., Z.W. and L.Y. designed the project. Material preparation and data collection and analysis were performed by Q.L. and Z.C. Y.T. and B.Z. measured SPR and inhibition assay. The first draft of the manuscript was written by Q.L. and Z.C. Q.W. and L.Y. checked and edited the manuscripts. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liao, Q., Chen, Z., Tao, Y. et al. An integrated method for optimized identification of effective natural inhibitors against SARS-CoV-2 3CLpro. Sci Rep 11, 22796 (2021). https://doi.org/10.1038/s41598-021-02266-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-02266-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.