Abstract
This paper presents label-free electrochemical transduction as a suitable scheme for COVID-19-specific viral RNA/c-DNA detection, with an aim to facilitate point of care diagnosis. In lieu of this, we discuss the proposed electrochemical biosensing scheme, based on electrodeposited gold nanoparticles as the transducing elements. Specific to this approach, here, the protocols associated with the immobilization of the single-stranded probe nucleotide on to the biosensor, have also been laid out. This paper also discusses the methods of electrochemical analysis, to be used for data acquisition and subsequent calibration, in relation to target analyte detection. Towards facilitating portable diagnosis, development of miniaturized sensors and their integration with readout units have also been discussed.
Similar content being viewed by others
Introduction
Development of portable DNA sensors is of significant medical importance, as they find direct application in infectious disease diagnosis (Boon et al. 2000; Kallioniemi et al. 1994; Alwine et al. 1977). For example, using DNA sensors, in case of viral infections, the genetic material of the virus (be in the RNA, or the c-DNA) inside an infected host can be detected using a complementary capture probe. Further, such devices can be utilized for mutation detection, where the target genetic material can be a mutant DNA, and the capturing probe can be complement of it. Notably, DNA sensors capable of mutation detection can find application in cancerous mutation detection. Towards the development of such sensors, several transduction principles have been used in the past, by researchers, globally (Sassolas et al. 2008; Kim et al. 2004; Yan et al. 2016; Green and Norton 2015; Wang and Li 2011). Among them, electrochemical methods have been predominately explored, on account of their inherent advantages, in terms of sensitivity, limiting detection and amenability to miniaturization (Drummond et al. 2003; Akhavan et al. 2012; Ricci et al. 2012; Bakker and Telting-Diaz 2002). In general, solid state electrochemical DNA sensors use capture probe modified working electrodes, as the sensing interface, on to which the complementary target nucleotide is hybridized. The process of hybridization is then transduced in to a physically measurable entity, using one of the many sensitive electrochemical techniques such as electrochemical impedance spectroscopy, cyclic voltammetry, amperometry and differential pulse voltammetry. Further, electrochemical methods often facilitate label-free DNA detection, which does not involve any sandwiched assay, or additional signaling molecule other than the probe and target nucleotides. Such advantage is crucial, as it greatly simplifies the protocols and the chemistry associated with the development of the biosensors.
Keeping these in mind, in the past, we had adhered to label-free electrochemical sensing of DNA hybridization, for both wild type and mutant targets (Tripathy et al. 2017, 2018a, b, 2019). In addition, our group has developed a number of electrochemical immunoassays towards label-free detection of malarial parasite, atrazine and cardiac biomarkers (Paul et al. 2016; Supraja et al. 2019a, b, c). During the course of development of such devices, the associated protocols were optimized for achieving superior sensitivity. In this paper, we propose to use this well-established electrochemical scheme as a potential approach for COVID-19 diagnosis. Given the current global situation, in relation to the ongoing pandemic, it is obviously essential that diagnosis methods are developed that are rapid, sensitive, target-specific, reproducible, technically robust, and cost effective. In particular, in case of the envisaged diagnosis platform, two important aspects are worth mentioning, when one considers using them in the field. The first point that needs attention is the portability of these sensors, which would allow them to be used as point-of-care diagnostic devices. Note, the problem of portability of these devices include the issue of device miniaturization, their interfacing with read-out systems, and subsequent data acquisition. Secondly, the economic aspects of a diagnostic device are of concern. Ideally, the cost associated with a diagnostic tool, which is to be used for mass-testing, has to be limited to affordable figures. Keeping both these aspects in mind, in this paper, an electrochemical DNA sensing approach has been discussed for the specific application.
We had earlier reported about electrochemical detection of Dengue virus specific single-stranded consensus primer, both from spiked buffer and spiked serum sample, using Mn2O3 nanofibers as transducing elements (Tripathy et al. 2017).Therein, the sensing interface was prepared via dropcasting of the Mn2O3 nanofibers on to commercial available glassy carbon electrodes (GCEs), and subsequently, chemically anchoring the probe nucleotides onto the nanofibers. At a later stage, we had also developed another electrochemical platform using GCEs and graphene doped Mn2O3 nanofibers, for the detection of BRCA-1 gene specific wild type and mutant DNAs (Tripathy et al. 2018a). However, in both these cases, the DNA sensors were macroscopic. Their utilization in on-field diagnosis, understandably, needed miniaturization of the sensing platform and its integration with a portable readout system. To achieve this, we had proposed a novel fabrication protocol for developing a miniaturized electrochemical biosensor, and demonstrated the usability of the device towards DNA hybridization and point mutation detection, using PCR-amplified samples (Tripathy et al. 2019). The concerned platform was envisaged to be a portable DNA-sensor, that can be used towards infectious disease diagnosis, such as Dengue and other viral or bacterial infections. In this paper, we intend to propose a plausible extension of the same platform towards COVID-19 diagnosis. The subsequent sections of this paper describe the detailed methodology and protocols associated with the same.
Methods and Protocols
Device Development
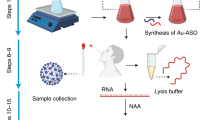
The proposed miniaturized device was developed on oxidized silicon substrates, using standard CMOS fabrication process flow and electrodeposition techniques. The working electrode for the sensor was realized with gold nanoparticles electrodeposited on to a Ti electrode, following previously established protocols (Tripathy et al. 2018b; Yi and Yu 2009). The electrodeposition conditions were experimentally optimized, for desirable performance. The reference and counter electrodes were of Platinum. Here, for external interfacing of the biosensor, a male USB port was attached on to the platform, using an indigenous strategy. Further, a PDMS reservoir was anchored on to the device, which defined the reaction area, and effectively ensured confinement of the electrodes. A detailed description of the process flow has been provided elsewhere (Tripathy et al. 2019). In Fig. 1, a schematic representation of the device is presented. Following the same protocol, a handheld electrochemical platform, with a well-defined reaction chamber, and a USB-based interfacing arrangement, can be developed, for COVID-19 specific DNA sensing. In addition, however, one may also opt for other fabrication techniques for developing the platform. Note, cost-effective methods such as screen printing of conductive metal inks on to low-cost platforms can be considered as alternatives.
Protocols for Developing the Sensing Interface
The proposed electrochemical platform makes use of gold nanoparticles, electrodeposited on to a Ti surface, as the sensing electrode. This comes as a strategic improvement, in terms of stability and simplicity, over sensing interfaces that are prepared by physisorption of nanomaterials on to solid surfaces. Such surfaces, in general, are prone to inter-device variability, and are not wash proof. They stability of such interfaces against wet chemical steps is a matter of concern, as it may induce unwanted ‘leaching’ of material. On the contrary, the electrodeposited Au layer is highly stable, and immune to harsh chemical treatments, Au being a noble metal. This improvement in stability is desirable for on-field biomolecular detection, in relation to accuracy and reproducibility. To add to this, the Au nanostructure-based sensing interface is advantageous in terms of simplicity of biofunctionalization, as it facilitates the use of gold-thiol self-assembly method. Such a protocol is pretty well understood, and highly followed for DNA immobilization (Lucarelli et al. 2004; Zhang et al. 2007; Rasheed and Sandhyarani 2017).
For the concerned case of COVID-19 diagnosis, this protocol can be utilized as shown in Fig. 2. Here, the target nucleotide can be the COVID-19 specific viral RNA, or the corresponding c-DNA, or any unique sequence specific to them. A complementary single-stranded probe can then be designed, with respect to the target sequence, with thiol modification at one end. The thiol modified probe can be attached on to the gold sensing electrodes via gold-thiol self-assembly, as described before. And subsequently, the sensing surface can be made diagnosis-ready, by blocking the non-specific binding sites, using any standard blocking agent. When the target nucleotide is introduced on to the sensor, through the designated reaction chamber, it hybridizes with the complementary probe, under favorable physiological conditions. This hybridization can be recorded using electrochemical techniques as described in the next sequence, and thereby, a diagnosis scheme is developed.
Data Acquisition and Calibration
As discussed previously, the target hybridization on the sensor surface can be recorded via several electrochemical techniques, that converts this biophysical binding event in to a proportional electrical signal. A great deal of literature can be found in relation to these methods, and their advantages and shortcomings. Here, we are not discussing them in details. In this paper, we only intend to provide a means of data acquisition, that can be used in conjunction to the developed miniaturized platform, for COVID-19-specific diagnosis.
In our previous publications, we had used differential pulse voltammetry and electrochemical impedance spectroscopy, for recording the electrochemical response of DNA sensors (Tripathy et al. 2017, 2018a,b, 2019). Either of these methods can be used in the current situation, and in addition to them, one may also opt for the comparatively simpler method of amperometry. In either of the cases, an electrical signal (be it current, voltage or impedance) is recorded, that depicts the change in reaction kinetics at the sensing interface, directly correlating the same to the concentration of the COVID-19-specific target DNA. In general, when the target DNA binds to the surface immobilized capture probe, and the hybridization occurs, the overall surface charge on the biosensor changes significantly, in proportion to the concentration of the bound target, for a given probe density. This change in surface charge, in turn, alters the reaction kinetics, which is subsequently reflected in the system response. The sensor response can be recorded using an electrochemical workstation or any equivalent readout. In our case, the proposed miniaturized platform can be interfaced with such workstations or readouts, through the USB interface. As a matter of fact, the platform can also be interfaced with PCB based readouts or Arduino boards, and the data acquisition can be performed on a desktop or laptop. In our previous work, we had demonstrated one such instance with respect to the proposed platform, where amperometric data was recorded using an Arduino UNO board, and the PLX-DAQv2.xlsm worksheet which contains a macro (Tripathy et al. 2019). The arrangement, however, can be extended to other methods, for the COVID-19 specific diagnosis.
Also, towards ensuring point-of-care diagnosis, it is imperative that portable read-out platforms are developed, that are simple and user-friendly. New and portable systems, in essence, can get rid of the bulky and table top electrochemical workstations, such that the sensors can be operated on field, without a laboratory facility. For a smarter healthcare, one can ideally aim at developing a readout that can be interfaced with smartphones and operated via software applications. In Fig. 3, a pictorial schematic is presented, showing the envisaged interfacing of the miniaturized device with a smartphone. Such systems will definitely increase the impact of diagnosis kits by manifolds, as with their availability, point-of-care biosensing can be performed on a larger scale, and the benefits of healthcare can reach a larger community.
Conclusion
In this paper, label-free electrochemical detection of DNA hybridization has been presented as a potential approach for COVID-19 diagnosis. In relation to this, fabrication process flow of a miniaturized electrochemical biosensor is reported, and the related protocols for DNA sensing are laid out. As described, using complementary thiolated probes, COVID-19 specific viral RNA or c-DNA can be detected, and quantified, with the proposed platform. Note, development of such a full-fledged device may take 3–6 months, to be used in the concerned diagnosis. However, the DNA-hybridization based disease diagnosis requires the extraction of target DNA/RNA from the infected host and the subsequent sample preparation. Additionally, in this paper, we have presented a roadmap for interfacing the proposed device with smartphone based read-out systems, for smart healthcare. Such a system would eliminate the need of using bulky instrumentation for data acquisition from electrochemical sensors, and facilitate point-of-care diagnosis.
References
Akhavan O, Ghaderi E, Rahighi R (2012) Toward single-DNA electrochemical biosensing by graphene nanowalls. ACS Nano 6(4):2904–2916
Alwine JC, Kemp DJ, Stark GR (1977) Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci 74(12):5350–5354
Bakker E, Telting-Diaz M (2002) Electrochemical sensors. Anal Chem 74(12):2781–2800
Boon EM, Ceres DM, Drummond TG, Hill MG, Barton JK (2000) Mutation detection by electrocatalysis at DNA-modified electrodes. Nat Biotechnol 18(10):1096–1100
Drummond TG, Hill MG, Barton JK (2003) Electrochemical DNA sensors. Nat Biotechnol 21(10):1192
Green NS, Norton ML (2015) Interactions of DNA with graphene and sensing applications of graphene field-effect transistor devices: a review. Anal Chim Acta 853:127–142
Kallioniemi OP, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D (1994) Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosom Cancer 10(4):231–243
Kim DS, Jeong YT, Park HJ, Shin JK, Choi P, Lee JH, Lim G (2004) An FET-type charge sensor for highly sensitive detection of DNA sequence. Biosens Bioelectron 20(1):69–74
Lucarelli F, Marrazza G, Turner AP, Mascini M (2004) Carbon and gold electrodes as electrochemical transducers for DNA hybridisation sensors. Biosens Bioelectron 19(6):515–530
Paul KB, Kumar S, Tripathy S, Vanjari SRK, Singh V, Singh SG (2016) A highly sensitive self-assembled monolayer modified copper doped zinc oxide nanofiber interface for detection of Plasmodium falciparum histidine-rich protein-2: Targeted towards rapid, early diagnosis of malaria. Biosens Bioelectron 80:39–46
Rasheed PA, Sandhyarani N (2017) Electrochemical DNA sensors based on the use of gold nanoparticles: a review on recent developments. Microchim Acta 184(4):981–1000
Ricci F, Adornetto G, Palleschi G (2012) A review of experimental aspects of electrochemical immunosensors. Electrochim Acta 84:74–83
Sassolas A, Leca-Bouvier BD, Blum LJ (2008) DNA biosensors and microarrays. Chem Rev 108(1):109–139
Supraja P, Sudarshan V, Tripathy S, Agrawal A, Singh SG (2019a) Label free electrochemical detection of cardiac biomarker troponin T using ZnSnO3 perovskite nanomaterials. Anal Meth 11(6):744–751
Supraja P, Tripathy S, Vanjari SRK, Singh V, Singh SG (2019b) Label free, electrochemical detection of atrazine using electrospun Mn2O3 nanofibers: towards ultrasensitive small molecule detection. Sens Actuators B Chem 285:317–325
Supraja P, Tripathy S, Vanjari SRK, Singh V, Singh SG (2019c) Electrospun tin (IV) oxide nanofiber based electrochemical sensor for ultra-sensitive and selective detection of atrazine in water at trace levels. Biosens Bioelectron 141:111441
Tripathy S, Vanjari SRK, Singh V, Swaminathan S, Singh SG (2017) Electrospun manganese (III) oxide nanofiber based electrochemical DNA-nanobiosensor for zeptomolar detection of dengue consensus primer. Biosens Bioelectron 90:378–387
Tripathy S, Gangwar R, Supraja P, Rao AN, Vanjari SRK, Singh SG (2018a) Graphene doped Mn2O3 nanofibers as a facile electroanalytical DNA point mutation detection platform for early diagnosis of breast/ovarian cancer. Electroanalysis 30(9):2110–2120
Tripathy S, Joseph J, Vanjari SRK, Rao AN, Singh SG (2018b) Flexible ITO electrode with gold nanostructures for femtomolar DNA hybridization detection. IEEE Sens Lett 2(4):1–4
Tripathy S, Joseph J, Pothuneedi S, Das D, Vanjari SRK, Rao AN, Singh SG (2019) A miniaturized electrochemical platform with an integrated PDMS reservoir for label-free DNA hybridization detection using nanostructured Au electrodes. Analyst 144(23):6953–6961
Wang L, Li PC (2011) Microfluidic DNA microarray analysis: a review. Anal Chim Acta 687(1):12–27
Yan Y, Samai S, Bischoff KL, Zhang J, Ginger DS (2016) Photocontrolled DNA hybridization stringency with fluorescence detection in heterogeneous assays. ACS Sens 1(5):566–571
Yi Q, Yu W (2009) Nanoporous gold particles modified titanium electrode for hydrazine oxidation. J Electroanal Chem 633(1):159–164
Zhang J, Song S, Wang L, Pan D, Fan C (2007) A gold nanoparticle-based chronocoulometric DNA sensor for amplified detection of DNA. Nat Protoc 2(11):2888
Acknowledgements
The authors acknowledge Department of science and technology (Fund for Improvement of S&T infrastructure), Government of India and Board of research in nuclear sciences (BRNS), Government of India for funding the research.
Funding
The authors acknowledge Department of science and technology (Fund for Improvement of S&T infrastructure), Government of India and Board of research in nuclear sciences (BRNS), Government of India for funding the research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Availability of data and material (data transparency)
Not applicable.
Code availability (software application or custom code)
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tripathy, S., Singh, S.G. Label-Free Electrochemical Detection of DNA Hybridization: A Method for COVID-19 Diagnosis. Trans Indian Natl. Acad. Eng. 5, 205–209 (2020). https://doi.org/10.1007/s41403-020-00103-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41403-020-00103-z