ISSN: 0973-7510
E-ISSN: 2581-690X
Coronavirus disease – 2019 (COVID-19) pandemic, due to severe acute respiratory syndrome–coronavirus-2 (SARS-CoV-2), is posing a severe bio threat to the entire world. Nucleocapsids of SARS-CoV-2 and the related viruses were studied for gene and amino acid sequence homologies. In this study, we established similarities and differences in nucleocapsids in SARS-CoV-2, severe acute respiratory syndrome – coronavirus-1 (SARS-CoV-1), bat coronavirus (bat-CoV) and Middle East respiratory syndrome – coronavirus (MERS-CoV). We conducted a detailed analysis of the nucleocapsid protein amino acid and gene sequence encoding it, found in various coronavirus strains. After thoroughly screening the different nucleocapsids, we observed a close molecular homology between SARS-CoV-1 and SARS-CoV-2. More than 95% sequence similarity was observed between the two SARS-CoV strains. Bat-CoV and SARS-CoV-2 showed 92% sequence similarity. MERS-CoV and SARS-CoV-2 nucleocapsid analysis indicated only 65% identity. Molecular characterization of nucleocapsids from various coronaviruses revealed that SARS-CoV 2 is more related to SARS-CoV 1 and bat-CoV. SARS-CoV 2 exhibited less resemblance with MERS-CoV. SARS-CoV 2 showed less similarity to MERS-CoV. Thus, either SARS-CoV-1 or bat-CoV may be the source of SARS-CoV-2 evolution. Moreover, the existing differences in nucleocapsid molecular structures in SARS-CoV-2 make this virus more virulent and highly infectious, which means that the non-identical SARS-CoV-2 genes (which are absent in SARS-CoV-1 and bat-CoV) are responsible for COVID-19 severity. We observed that SARS-CoV-2 nucleocapsid from different locations varied in amino acid sequences. This revealed that there are many SARS-CoV-2 subtypes/subsets currently circulating globally. This study will help to develop antiviral vaccine and drugs, study viral replication and immunopathogenesis, and synthesize monoclonal antibodies that can be used for precise COVID-19 diagnosis, without false-positive/false-negative results.
SARS-CoV-2, SARS-CoV-1, Bat-CoV, MERS, Nucleocapsid (N) protein, COVID-19, Correlation, Virulence
Coronaviruses are a relatively large (125 nm), diverse group of RNA viruses infecting many mammals, amphibians, reptiles, and birds1,2. Coronavirus contains a positive-sense RNA (+ RNA) genome that directly acts as messenger RNA (mRNA) during viral replication inside the host cell3. Coronaviruses are spherical particles with a viral envelope, which is acquired from host cytoplasmic membrane4. Although coronaviruses are enveloped, the envelope is very sensitive to temperature, chemicals, disinfectants, soaps, and sanitizers5. The viral envelope is crucial for host cell attachment, leading to further replication steps6. If the viral envelope is disintegrated, coronaviruses cannot proceed with replication7. On the viral envelope surface, there are spike glycoproteins, which are very important for attachment to host cell receptors8. Severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) viral spike glycoproteins attach to angiotensin-converting enzyme-2 (ACE-2) receptors on pneumocytes9. ACE-2 receptors are located in the lungs, heart, arteries, kidneys, and intestines10. Recent case studies have revealed that many patients with coronavirus disease-2019 (COVID-19) have also died of myocardial infarction, stroke, and renal failure11–13. This suggests that after viremia, SARS-CoV-2, in addition to severe lung infection, can also infect other body sites. Inside the viral envelope, coronavirus possesses a nucleocapsid (N) protein, which is very important for its replication, antigenesis, pathogenesis, virulence, infection and, dissemination14. This N protein is helically symmetrical with RNA coiling around it15. All heredity information for viral spike protein synthesis, replication mechanism, other viral encoded protein synthesis, host cell docking and damage, and disease progression are encoded by the viral RNA in N protein16,17. Coronavirus N protein determines all aspects of viral pathogenesis. In our study, we attempted to characterize the N protein. The N proteins present in other coronaviruses were also compared during this study. We analyzed the molecular composition and amino acid sequence of N present in SARS-CoV-2 and other related viruses like SARS-CoV-1, bat-CoV, and Middle East respiratory syndrome- coronavirus (MERS-CoV). This will help virologists to study viral replication, viral-host immunopathogenesis, and vaccine development to combat the current COVID-19 pandemic. To date, the COVID-19 pandemic has resulted in more than 3,672,238 cases and 254,045 deaths worldwide (WHO-Situation Report-108). This figure is rapidly increasing worldwide, suggesting that SARS-CoV-2 is rapidly disseminating among communities. The rapid infection rate may be attributed to the low virus infectious dose required to elicit infection (0.01 PFU/ml) in an individual18. Reports suggest that temperature variation in different countries also plays an important role in the spread of COVID-1919,20. Other studies speculate that individuals already immunized with the BCG vaccine have protection against this disease21,22; however, there is no effective vaccine available for COVID-19. There are also no antiviral drugs available for COVID-19. Some scientists are empirically employing antimalarial drugs like hydroxychloroquine, and antibiotics such as azithromycin, for COVID-1923–25. The USA Food and Drug Administration (FDA) permitted the use of the antiviral drug remdesivir for COVID-19 patients. The actual antiviral effects of these drugs against COVID-19 have not been thoroughly proven26. In these circumstances, there is a need for a suitable killed/attenuated vaccine, which combats SARS-CoV-2, for immunoprophylaxis. If an effective COVID-19 vaccine is established, it will elicit a good immune response containing SARS-CoV-2 infection in the community after administration. To develop a vaccine and antiviral drugs against COVID-19, N protein molecular characterization is crucial. Therefore, in this study, we employed various bioinformatics tools, GenBank, and the European nucleotide archive (ENA) to compare SARS-CoV-2 N protein with other related coronaviruses. This study emphasizes and suggests ways to contain and combat COVID-19.
Data Sources and Research Strategies
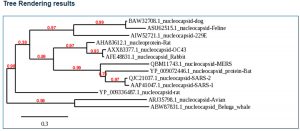
We employed bioinformatics tools and used data present in various gene banks, and amino acid sequences were analyzed using related amino acid sequence banks for specific proteins. This research concentrated on GenBank (NLM-NIH) and the European Nucleotide Archive (ENA). We also employed bioinformatics tools like BLAST analysis of N protein. N proteins of various coronaviruses like SARS-CoV-1, bat-CoV, and MERS were compared with SARS-CoV-2 (COVID-19). After the analysis of N proteins of these coronaviruses, a phylogenetic tree was also established. Our research also carried out a thorough and detailed literature review on molecular aspects of coronaviruses, using available databases such as MEDLINE/PubMed, SCOPUS, Web of Science, ScienceDirect, and Google Scholar. We used combined MeSH (Medical Subject Headings) terms in Google Scholar such as ‘COVID-19’, ‘Coronavirus’, ‘SARS-CoV-2’, ‘SARS-CoV-1’, ‘bat-CoV’, ‘MERS-CoV’, ‘Nucleocapsid protein’, ‘N-Protein’ etc. for the search process from different databases.
Eligibility Criteria
In this study, SARS-CoV-2 was molecularly characterized. Its molecular composition was also compared with other coronaviruses such as SARS-CoV-1, bat-CoV, and MERS-CoV. For literature review, articles were included on the following basis: (a) majorly published in English between 2000 and 2020; (b) accounts of successful trials and experimentations on coronaviruses (c) recent WHO and CDC COVID-19 reports; (d) viral genomic studies (e) original and peer-reviewed articles (f) Coronavirus data available in GenBank and the ENA.
Articles were excluded on the following basis: (a) insufficient or no data; (b) not having a proper study design or approach.
Tools employed
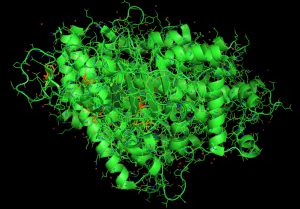
NCBI Genome WorkBench was employed for sequence processing and Clustal-X (Version 2.1) for sequence alignment. NCBI Tree Viewer was used to visualize the phylogenetic tree. Additionally, python tools were employed in JupyterLab Notebook (version 1.0) to explore SARS-CoV-2, SARS-CoV-1, bat-CoV and MERS-CoV N-protein structure, obtained from different hosts such as bats (Chiroptera), human (Homo sapiens) worldwide, rabbits (Oryctolagus cuniculus), brown rats (Rattus norvegicus), and many other possible potential SARS-Cov-2 hosts updated in NCBI till date. BioPython (version 1.76) was used for importing modules. We used PyMol software for molecular visualization of the SARS-CoV-2 N-protein structure. During this study, we employed GenBank (NLM-NIH) and ENA for gene and protein sequence analyses.
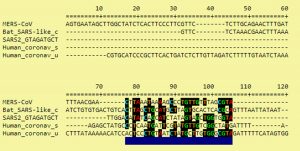
When the nucleocapsids of SARS-CoV-2, SARS-CoV-1, bat coronavirus, and MERS-CoV amino acid sequences were analyzed, we found 95% homology existing between SARS-CoV-2 and SARS-CoV-1 (Fig. 1, Fig. 2). SARS-CoV-2 and bat coronavirus nucleocapsids showed 92% amino acid sequence similarity (Fig. 1, Fig. 2). When N protein of SARS-CoV-2 and MERS-CoV were analyzed, only 65% amino acid sequence similarity was observed. Additional to the above mentioned non-COVID-19 viruses, we have also analyzed other beta coronaviruses with different animal origins with SARS-CoV-2. For this study, we used nucleocapsid sequences of whale, rat, rabbit, and fowl coronaviruses. After analysis, we observed 45 to 55% N protein gene sequence homology between SARS-CoV-2 and these beta coronaviruses. In this study, we have established three dimensional (3D) structures of both SARS-CoV-2 (Fig. 3) and SARS-CoV-1 N proteins. This tertiary N protein structure of SARS-CoV-2 and SARS-CoV-1 indicated that these two structures, from two different virus clans, are more than 95% similar. When N proteins (3 D structures) were analyzed, we found that SARS-CoV-1 and bat coronavirus have close similarity (96%) with SARS-CoV-2. Surprisingly, we observed that SARS-CoV-2 nucleocapsids from Chinese and Italians are showing 1–2% variations. So, we speculated that SARS-CoV-2 might have its origin from SARS-CoV-1 or bat coronavirus. Otherwise, SARS-CoV-2 might have evolved from other SARS-related unidentified animal coronaviruses. Our study indicated that nucleocapsids of examined animal coronaviruses are not closely related to SARS-CoV-2. This is also confirmed by the 3D structure comparison with analyzed coronaviruses.
Many researchers observed that there is SARS-CoV-2 reinfection in the same treated patients27–29. In other cases and parts of the world, COVID-19 patients exhibited renal failure, myocardial infarction, diarrhea, and stroke30-34. The COVID-19 pattern in different countries showed varying severities35-37. Many patients are asymptomatic, yet capable of transmitting the infection to others38-41. Clinical features of COVID-19 in different parts of the world are also showing variations42-45. We observed that these variations in severity, clinical features, and reinfection of SARS-CoV-2 may be attributed to the ability of the virus to mutate. In our study, we found that SARS-CoV-2 identified in different geographical locations show little variations. We speculated that this variation may be responsible for the strain/clan/subset variations. We also presumed in our study that there may be many SARS-CoV-2 subsets circulating the world. These subsets may show different virulence, pathogenicity, and disease patterns.
SARS-CoV-2 and SARS-CoV-1 are closely related coronaviruses, whereas MERS-CoV is less similar to SARS-CoV-2. In our analysis, we observed that bat coronavirus and some animal coronaviruses have less N homology with SARS-CoV-2. N protein is very important for virus virulence, antigenicity, pathogenicity, and clinical severity. SARS-CoV-2 may have evolved from SARS-CoV-1. We also observed that there is variation between SARS-CoV-2 identified in different parts of the world. This suggests that SARS-CoV-2 has many subsets circulating in the world.
ACKNOWLEDGMENTS
We would like acknowledge the faculty members of School of Medicine, The Maldives National University for their critical reviews, comments and suggestions during this manuscript preparation.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
- Kannan S, Shaik Syed Ali P, Sheeza A, Hemalatha K. COVID-19 (Novel Coronavirus 2019) – recent trends. Eur Rev Med Pharmacol Sci. 2020;24(4):2006-2011.
- Culp WC. Coronavirus disease 2019. A A Pract. 2020;14(6):e01218.
Crossref - Gralinski LE, Menachery VD. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12(2):pii: E135.
Crossref - Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;
Crossref - Lu X, Zhang L, Du H, et al. SARS-CoV-2 infection in children. N Engl J Med. 2020;382:1663-1665.
Crossref - Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875-879.
Crossref - Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281-292. e6.
Crossref - Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;
Crossref - Tai W, He L, Zhang X, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;
Crossref - Han Y, Kral P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14(4):5143-5147.
Crossref - Li B, Yang J, Zhao F, et al. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin Res Cardiol. 2020;109:531-538.
Crossref - Zhu ZW, Tang JJ, Chai XP, et al. Comparison of heart failure and 2019 novel coronavirus pneumonia in chest CT features and clinical characteristics. Zhonghua Xin Xue Guan Bing Za Zhi. 2020;48(0):E007.
- Chen L, Li X, Chen M, Feng Y, Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc Res. 2020;116(6):1097-1100.
Crossref - Kim JM, Chung YS, Jo HJ, et al. Identification of coronavirus isolated from a patient in Korea with Covid-19. Osong Public Heal Res Perspect. 2020;11(1):3-7.
Crossref - Xu X, Yu C, Qu J, et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur J Nucl Med Mol Imaging. 2020;47:1275-1280.
Crossref - Jin Y, Yang H, Ji W, et al. Virology, epidemiology, pathogenesis, and control of Covid-19. Viruses. 2020;12(4):372.
Crossref - Yi Y, Lagniton PNP, Ye S, Li E, Xu RH. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int J Biol Sci. 2020;16(10):1753-1766.
Crossref - Pedersen SF, Ho YC. SARS-CoV-2: a storm is raging. J Clin Invest. 2020;130(5):2202-2205.
Crossref - Sajadi MM, Habibzadeh P, Vintzileos A, Shokouhi S, Miralles-Wilhelm F, Amoroso A. Temperature and latitude analysis to predict potential spread and seasonality for COVID-19. SSRN Electron J. 2020.
Crossref - Xu K, Chen Y, Yuan J, et al. Factors associated with prolonged viral RNA shedding in patients with COVID-19. Clin Infect Dis. 2020; pii: ciaa351.
Crossref - Sala G, Miyakawa T. Association of BCG vaccination policy with prevalence and mortality of COVID-19. medRxiv. 2020.
Crossref - Szigeti R, Kellermayer D, Kellermayer R. BCG protects against COVID-19? A word of caution. medRxiv. 2020.
Crossref - Touret F, de Lamballerie X. Of chloroquine and COVID-19. Antiv Res. 2020;
Crossref - Gautret P, Lagier J-C, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020.
Crossref - Cortegiani A, Ingoglia G, Ippolito M, Giarratano A, Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care. 2020; pii: S0883-9441(20)30390-7.
Crossref - Zhai P, Ding Y, Wu X, Long J, Zhong Y, Li Y. The epidemiology, diagnosis and treatment of COVID-19. Int J Antimicrob Agents. 2020.
Crossref - Luo A. Positive SARS-CoV-2 test in a woman with COVID-19 at 22 days after hospital discharge: A case report. J Tradit Chinese Med Sci. 2020.
Crossref - Zhang J Jin, Dong X, Cao Y Yuan, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy Eur J Allergy Clin Immunol. 2020.
Crossref - Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis. 2020;10(2):102-108.
Crossref - Wong SH, Lui RN, Sung JJ. Covid-19 and the digestive system. J Gastroenterol Hepatol. 2020;35(5):744-748.
Crossref - D’Amico F, Baumgart DC, Danese S, Peyrin-Biroulet L. Diarrhea during COVID-19 infection: pathogenesis, epidemiology, prevention and management. Clin Gastroenterol Hepatol. 2020.
Crossref - Cheung KS, Hung IF, Chan PP, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology. 2020.
Crossref - South AM, Diz D, Chappell MC. COVID-19, ACE2 and the cardiovascular consequences. Am J Physiol Heart Circul Physiol. 2020;318(5):H1084-H1090.
Crossref - Han H, Xie L, Liu R, et al. Analysis of heart injury laboratory parameters in 273 COVID-19 patients in one hospital in Wuhan, China. J Med Virol. 2020.
Crossref - Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;16(24):91-98.
Crossref - Li Y C, Bai WZ, Hashikawa T. The neuroinvasive potential of SARS-CoV2 may be at least partially responsible for the respiratory failure of COVID-19 patients. J Med Virol. 2020.
Crossref - Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrobial Agents. 2020;55(3):105924.
Crossref - Lai CC, Liu YH, Wang CY, et al. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. J Microbiol Immunol Inf. 2020; pii: S1684-1182(20)30040-2.
Crossref - Kimball A, Hatfield KM, Arons M, et al. Asymptomatic and presymptomatic SARS-COV-2 infections in residents of a long-term care skilled nursing facility – King County, Washington, March 2020. MMWR. 2020;69(13):377-381.
Crossref - Zhang JF, Yan K, Ye HH, Lin J, Zheng JJ, Cai T. SARS-CoV-2 turned positive in a discharged patient with COVID-19 arouses concern regarding the present standard for discharge. Int J Infect Dis. 2020; pii: S1201-9712(20)30126-0.
Crossref - Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. 2020.
Crossref - Lake MA. What we know so far: COVID-19 current clinical knowledge and research. Clin Med. 2020;20(2):124-127.
Crossref - Wu D, Wu T, Liu Q, Yang Z. The SARS-CoV-2 outbreak: What we know. Int J Infect Dis. 2020;94:44-48.
Crossref - Chen H, Guo J, Wang C, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020.
- Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):572-582.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.