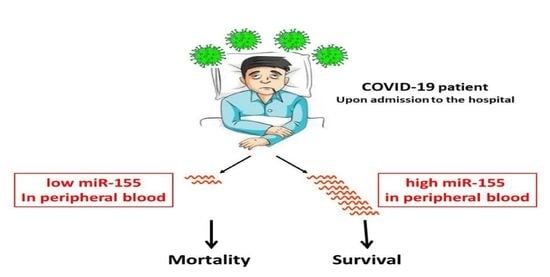
miR-155: A Potential Biomarker for Predicting Mortality in COVID-19 Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Patients
2.3. Data Collection
2.4. Sample Preparation, RNA Extraction, and cDNA Preparation and Quantification
2.5. Statistical Analysis
3. Results
3.1. miR-155 and miR-146b Are Differently Expressed in COVID-19 Patients
3.2. miR-155 Predicts Patient Outcome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grasselli, G.; Greco, M.; Zanella, A.; Albano, G.; Antonelli, M.; Bellani, G.; Bonanomi, E.; Cabrini, L.; Carlesso, E.; Castelli, G.; et al. Risk Factors Associated With Mortality Among Patients With COVID-19 in Intensive Care Units in Lombardy, Italy. JAMA Intern. Med. 2020, 180, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Ponti, G.; Maccaferri, M.; Ruini, C.; Tomasi, A.; Ozben, T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab. Sci. 2020, 57, 389–399. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandao, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Rice, T.W.; Wheeler, A.P.; Bernard, G.R.; Hayden, D.L.; Schoenfeld, D.A.; Ware, L.B. National Institutes of Health, N.H.L.; Blood Institute, A.N., Comparison of the SpO2/FIO2 ratio and the PaO2/FIO2 ratio in patients with acute lung injury or ARDS. Chest 2007, 132, 410–417. [Google Scholar] [CrossRef]
- Lakens, D. Calculating and reporting effect sizes to facilitate cumulative science: A practical primer for t-tests and ANOVAs. Front. Psychol. 2013, 4, 863. [Google Scholar] [CrossRef] [Green Version]
- Wong, R.S.Y. Inflammation in COVID-19: From pathogenesis to treatment. Int. J. Clin. Exp. Pathol. 2021, 14, 831–844. [Google Scholar]
- Mann, M.; Mehta, A.; Zhao, J.L.; Lee, K.; Marinov, G.K.; Garcia-Flores, Y.; Lu, L.F.; Rudensky, A.Y.; Baltimore, D. Author Correction: An NF-kappaB-microRNA regulatory network tunes macrophage inflammatory responses. Nat. Commun. 2018, 9, 3338. [Google Scholar] [CrossRef]
- Comer, B.S.; Camoretti-Mercado, B.; Kogut, P.C.; Halayko, A.J.; Solway, J.; Gerthoffer, W.T. MicroRNA-146a and microRNA-146b expression and anti-inflammatory function in human airway smooth muscle. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 307, L727–L734. [Google Scholar] [CrossRef]
- Lu, T.X.; Munitz, A.; Rothenberg, M.E. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 2009, 182, 4994–5002. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Li, J.; Yao, H.; Li, T. Restoring microRNA-499-5p Protects Sepsis-Induced Lung Injury Mice Via Targeting Sox6. Nanoscale Res. Lett. 2021, 16, 89. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, X.P.; Li, Y.J. MicroRNA-146a and human disease. Scand. J. Immunol. 2010, 71, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, H.; Yang, B. miR-146a regulates inflammation and development in patients with abdominal aortic aneurysms by targeting CARD10. Int. Angiol. A J. Int. Union Angiol. 2020, 39, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, C.; Gu, W.; Ji, H.; Zhu, L. Serum Exosomal MicroRNAs Predict Acute Respiratory Distress Syndrome Events in Patients with Severe Community-Acquired Pneumonia. BioMed Res. Int. 2019, 2019, 3612020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, L.F.; Gasteiger, G.; Yu, I.S.; Chaudhry, A.; Hsin, J.P.; Lu, Y.; Bos, P.D.; Lin, L.L.; Zawislak, C.L.; Cho, S.; et al. A Single miRNA-mRNA Interaction Affects the Immune Response in a Context- and Cell-Type-Specific Manner. Immunity 2015, 43, 52–64. [Google Scholar] [CrossRef] [Green Version]
- Backes, S.; Shapiro, J.S.; Sabin, L.R.; Pham, A.M.; Reyes, I.; Moss, B.; Cherry, S.; tenOever, B.R. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe 2012, 12, 200–210. [Google Scholar] [CrossRef] [Green Version]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef] [Green Version]
- Bala, S.; Csak, T.; Momen-Heravi, F.; Lippai, D.; Kodys, K.; Catalano, D.; Satishchandran, A.; Ambros, V.; Szabo, G. Biodistribution and function of extracellular miRNA-155 in mice. Sci. Rep. 2015, 5, 10721. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Sun, R.; Zhao, H.J.; Fu, D.; Zhong, H.J.; Weng, X.Q.; Qu, B.; Zhao, Y.; Wang, L.; Zhao, W.L. MiR155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+T cells. Mol. Cancer 2019, 18, 54. [Google Scholar] [CrossRef]
- Faccini, J.; Ruidavets, J.B.; Cordelier, P.; Martins, F.; Maoret, J.J.; Bongard, V.; Ferrieres, J.; Roncalli, J.; Elbaz, M.; Vindis, C. Circulating miR-155, miR-145 and let-7c as diagnostic biomarkers of the coronary artery disease. Sci. Rep. 2017, 7, 42916. [Google Scholar] [CrossRef] [Green Version]
- Garg, A.; Seeliger, B.; Derda, A.A.; Xiao, K.; Gietz, A.; Scherf, K.; Sonnenschein, K.; Pink, I.; Hoeper, M.M.; Welte, T.; et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur. J. Heart Fail. 2021, 23, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Edelman, J.J.; Kao, S.C.; Vallely, M.P.; van Zandwijk, N.; Reid, G. The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front. Genet. 2013, 4, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontecorvi, G.; Bellenghi, M.; Ortona, E.; Care, A. microRNAs as new possible actors in gender disparities of COVID-19 pandemic. Acta Physiol. 2020, 230, e13538. [Google Scholar] [CrossRef] [PubMed]
- Tacke, F.; Spehlmann, M.E.; Vucur, M.; Benz, F.; Luedde, M.; Cardenas, D.V.; Roy, S.; Loosen, S.; Hippe, H.J.; Frey, N.; et al. miR-155 Predicts Long-Term Mortality in Critically Ill Patients Younger than 65 Years. Mediat. Inflamm. 2019, 2019, 6714080. [Google Scholar] [CrossRef] [PubMed]
- Ong, J.; Woldhuis, R.R.; Boudewijn, I.M.; van den Berg, A.; Kluiver, J.; Kok, K.; Terpstra, M.M.; Guryev, V.; de Vries, M.; Vermeulen, C.J.; et al. Age-related gene and miRNA expression changes in airways of healthy individuals. Sci. Rep. 2019, 9, 3765. [Google Scholar] [CrossRef] [Green Version]
- Malik, P.; Patel, U.; Mehta, D.; Patel, N.; Kelkar, R.; Akrmah, M.; Gabrilove, J.L.; Sacks, H. Biomarkers and outcomes of COVID-19 hospitalisations: Systematic review and meta-analysis. BMJ Evid.-Based Med. 2021, 26, 107–108. [Google Scholar] [CrossRef]
- Loomba, R.S.; Villarreal, E.G.; Farias, J.S.; Aggarwal, G.; Aggarwal, S.; Flores, S. Serum biomarkers for prediction of mortality in patients with COVID-19. Ann. Clin. Biochem. 2021, 59, 15–22. [Google Scholar] [CrossRef]
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.O.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef]
- Li, C.; Hu, X.; Li, L.; Li, J.H. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J. Clin. Lab. Anal. 2020, 34, e23590. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Benitez, I.D.; Pinilla, L.; Carratala, A.; Moncusi-Moix, A.; Gort-Paniello, C.; Molinero, M.; Gonzalez, J.; Torres, G.; Bernal, M.; et al. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. J. Lab. Clin. Med. 2021, 236, 147–159. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Hijazi, Z.; Andersson, U.; Al-Khatib, S.M.; Lopes, R.D.; Alexander, J.H.; Held, C.; Hylek, E.M.; Leonardi, S.; Hanna, M.; et al. Use of Biomarkers to Predict Specific Causes of Death in Patients With Atrial Fibrillation. Circulation 2018, 138, 1666–1676. [Google Scholar] [CrossRef] [PubMed]
- Pregernig, A.; Muller, M.; Held, U.; Beck-Schimmer, B. Prediction of mortality in adult patients with sepsis using six biomarkers: A systematic review and meta-analysis. Ann. Intensive Care 2019, 9, 125. [Google Scholar] [CrossRef] [PubMed]



| Parameter | Healthy (n = 15) | COVID-19-Mild (n = 22) | COVID-19-Severe (n = 15) | p Value |
|---|---|---|---|---|
| Age (years) (Median (IQR)) | 46 (42–57) | 77.5 (62.25–88) | 55 (48–66) | <0.001 |
| Gender (male) | 4 (27%) | 14 (63.63%) | 12 (80%) | 0.003 |
| BMI | 25.0 (21.6–26.8) | 26 (25.1–29.33) | 31.5 (28.05–38.5) | 0.005 |
| Blood type n (%) | A+ 7 (47%) B+ 3 (20%) O+ 2 (13%) O− 2 (13%) N/A 1 (7%) | A+ 7 (31%) B+ 1 (4.5%) AB+ 3 (14%) O+ 2 (9%) N/A 9 (41%) | A+ 6 (40%) B+ 3 (20%) AB+ 1 (7%) O+ 3 (20%) N/A 2 (13%) | 0.35 |
| Diabetes | 1 (6.7%) | 8 (36.36%) | 1 (6.66%) | 0.25 |
| IHD | 0 (0) | 3 (13.63%) | 0 (0%) | 0.09 |
| CVE | 0 (0) | 2 (9.09%) | 1 (6.66%) | 0.63 |
| Malignancy | 0 (0) | 1 (4.54%) | 1 (6.66%) | 0.14 |
| HTN | 1 (6.7%) | 14 (63.63%) | 3 (20%) | 0.002 |
| Dyslipidemia | 1 (6.7%) | 9 (40.9%) | 3 (20%) | 0.046 |
| CRF | 0 (0) | 3 (13.63%) | 1 (6.66%) | 0.43 |
| COPD/CLD | 0 (0) | 3 (13.63%) | 0 (0%) | 0.38 |
| Thyroid disease | 0 (0) | 2 (9.09%) | 1 (6.66%) | 0.43 |
| Parameter | Mild (n = 22) | Severe (n = 15) | p Value |
|---|---|---|---|
| WBC peak (K/ μL) | 12.5 (10.06–16.27) | 17.4 (14.69–24.24) | 0.009 |
| HGB nadir (g/dL) | 11.84 (8.75–12.48) | 7.79 (7.09–10.89) | 0.004 |
| D-dimer peak (ng/mL) | 1262.5 (650.25–6469) | 16,724 (1158–28,955) | 0.01 |
| INR peak | 1.21 (1.03–1.35) | 1.39 (1.24–1.66) | 0.005 |
| Fibrinogen peak (mg/dL) | 541 (418–760.5) | 704 (601–886) | 0.04 |
| Creatinine peak (mg/dL) | 0.98 (0.78–1.48) | 1.18 (1.12–2.89) | 0.02 |
| Bilirubin peak (mg/dL) | 0.67 (0.51–0.74) | 1.28 (1.01–2.68) | 0.001 |
| AST-peak (IU/L) | 68.41 ± 45.62 | 810.6 ± 2544.42 | 0.17 |
| ALT-peak (IU/L) | 45.5 (25.5–99.75) | 161 (76–231) | <0.001 |
| LDH-peak (IU/L) | 445 (333.25–609.75) | 540 (515–1038) | 0.009 |
| Troponin-peak (ng/L) | 17.9 (8.4–28.77) | 38.2 (12.9–108) | 0.05 |
| CRP-peak (mg/L) | 146.5 (65.96–260.25) | 300 (234–356) | 0.004 |
| IL6 (pg/mL) (n = 16) | 53 (30.5–218.25) | 147.5 (20.75–336) | 0.38 |
| IL1B (pg/mL) (n = 16) | 1 (0.75–1) | 0 (0–1) | 0.20 |
| IL8 (pg/mL) (n = 16) | 110 (55.5–255) | 66 (44.25–118.25) | 0.27 |
| TNFα (pg/mL) (n = 16) | 32 (23–63) | 28.5 (16–42.5) | 0.55 |
| s/f ratio upon admission median (IQR) | 442.85 (328.68–447.61) | 146.66 (108.88–192) | <0.001 |
| Intubated (Yes) n (%) | 6 (27.27%) | 12 (80%) | 0.003 |
| Ventilation days n (%) | 0 (0–11.25) | 16 (3–33) | 0.006 |
| LOS-Hospitalized days n (IQR) | 14 (6–28) | 32 (21–50) | 0.01 |
| Mortality | 3 (13.63%) | 3 (20%) | 0.13 |
| ECMO | 0 (0%) | 4 (26.67%) | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kassif-Lerner, R.; Zloto, K.; Rubin, N.; Asraf, K.; Doolman, R.; Paret, G.; Nevo-Caspi, Y. miR-155: A Potential Biomarker for Predicting Mortality in COVID-19 Patients. J. Pers. Med. 2022, 12, 324. https://doi.org/10.3390/jpm12020324
Kassif-Lerner R, Zloto K, Rubin N, Asraf K, Doolman R, Paret G, Nevo-Caspi Y. miR-155: A Potential Biomarker for Predicting Mortality in COVID-19 Patients. Journal of Personalized Medicine. 2022; 12(2):324. https://doi.org/10.3390/jpm12020324
Chicago/Turabian StyleKassif-Lerner, Reut, Keren Zloto, Nadav Rubin, Keren Asraf, Ram Doolman, Gidi Paret, and Yael Nevo-Caspi. 2022. "miR-155: A Potential Biomarker for Predicting Mortality in COVID-19 Patients" Journal of Personalized Medicine 12, no. 2: 324. https://doi.org/10.3390/jpm12020324