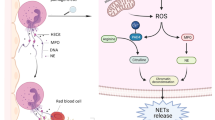
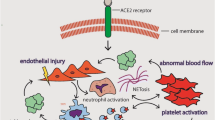
Abstract
Studies of patients with COVID-19 have demonstrated markedly dysregulated coagulation and a high risk of morbid arterial and venous thrombotic events. Elevated levels of blood neutrophils and neutrophil extracellular traps (NETs) have recently been described in patients with COVID-19. However, their potential role in COVID-19-associated thrombosis remains incompletely understood. In order to elucidate the potential role of hyperactive neutrophils and NET release in COVID-19-associated thrombosis, we conducted a case–control study of patients hospitalized with COVID-19 who developed thrombosis, as compared with gender- and age-matched COVID-19 patients without clinical thrombosis. We found that remnants of NETs (cell-free DNA, myeloperoxidase-DNA complexes, and citrullinated histone H3) and neutrophil-derived S100A8/A9 (calprotectin) in patient sera were associated with higher risk of morbid thrombotic events in spite of prophylactic anticoagulation. These observations underscore the need for urgent investigation into the potential relationship between NETs and unrelenting thrombosis in COVID-19, as well as novel approaches for thrombosis prevention.
Similar content being viewed by others
Highlights
-
Mechanisms contributing to frequent thrombosis in COVID-19 remain incompletely understood
-
NETs and neutrophil activation were measured in patients with COVID-19-associated thrombosis
-
Thrombosis in COVID-19 was associated with higher levels of circulating NETs and calprotectin
-
Exploring approaches to neutralize neutrophils and NETs in COVID-19 warrants urgent investigation
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes the disease known as coronavirus disease 2019 (COVID-19). It most commonly presents with influenza-like illness and viral pneumonia, but in its most severe manifestation progresses to acute respiratory distress syndrome (ARDS) and multi-organ failure [1]. To date the viral pandemic has resulted in millions of infections worldwide (https://coronavirus.jhu.edu/map.html).
In COVID-19, elevated levels of blood neutrophils predict severe respiratory disease and unfavorable outcomes [2, 3]. Neutrophil-derived extracellular traps (NETs) play a pathogenic role in many thrombo-inflammatory states including sepsis [4, 5], thrombosis [6,7,8], and respiratory failure [9, 10]. NETs are extracellular webs of chromatin and microbicidal proteins that are an evolutionarily conserved aspect of innate immune host-defense [11]; however, NETs also have potential to initiate and propagate inflammation and thrombosis. NETs deliver a variety of oxidant enzymes to the extracellular space, including myeloperoxidase, NADPH oxidase, and nitric oxide synthase, while also serving as a source of extracellular histones that carry significant cytotoxic potential. NETs drive cardiovascular disease by propagating inflammation in vessel walls [12]. Furthermore, when formed intravascularly, NETs can drive occlusion of arteries [13], veins [14], and microscopic vessels [15]. Studies of COVID-19 suggest a high risk of morbid arterial events [16], and the risk of venous thromboembolism (VTE) is increasingly revealing itself as more data become available [17].
Descriptive and mechanistic studies to date that examine COVID-19 pathophysiology have focused on monocytes and lymphocytes more so than neutrophils and their effector products [18]. Our group recently reported high levels of NETs in 50 patients hospitalized with COVID-19 as compared with healthy controls [19]. These findings have since been replicated by several other groups [19,20,21,22,23,25]. Here, we describe 11 cases of thrombosis in patients hospitalized with COVID-19 and demonstrate an association with neutrophil hyperactivity and NET release.
Methods
Human samples for NETs analysis
All 44 patients studied here had a confirmed COVID-19 diagnosis based on FDA-approved DiaSorin Molecular Simplexa COVID-19 Direct real-time RT-PCR assay. Blood was collected into serum separator tubes by a trained hospital phlebotomist. After completion of biochemical testing ordered by the clinician, the remaining serum was stored at 4 °C for 4 to 48 h before it was deemed “discarded” and released to the research laboratory. Serum samples were immediately divided into small aliquots and stored at − 80 °C until the time of testing. This study complied with all relevant ethical regulations, and was approved by the University of Michigan Institutional Review Board (HUM00179409), which waived the requirement for informed consent given the discarded nature of the samples.
Quantification of S100A8/A9 (calprotectin)
Calprotectin levels were measured with the Human S100A8/S100A9 Heterodimer DuoSet ELISA (DY8226-05, R&D Systems) according to the manufacturer’s instructions and as done previously [26]. DuoSet ELISA kits are designed to be development kits for research purpose only. The kits are tested for appropriate antibody pairing, reagent stability, parallelism using naturally-derived protein, cross-reactivity/interference of related proteins, and lot-to-lot variability. The kit is calibrated against a highly purified E. coli expressed recombinant human S100A8/A9 heterodimer produced at R&D. The limit of detection is 93.8 pg/ml. The expected coefficient of variance should be less than 10% when following manufacture’s recommendations and good laboratory practice.
Quantification of cell-free DNA
Cell-free DNA was quantified in sera using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, P11496) according to the manufacturer’s instructions.
Quantification of myeloperoxidase-DNA complexes
Myeloperoxidase-DNA complexes were quantified in sera similarly to what has been previously described [27, 28]. This protocol used several reagents from the Cell Death Detection ELISA kit (Roche). First, a high-binding EIA/RIA 96-well plate (Costar) was coated overnight at 4ºC with anti-human myeloperoxidase antibody (Bio-Rad 0400-0002), diluted to a concentration of 1 µg/ml in coating buffer (Cell Death kit). The plate was washed two times with wash buffer (0.05% Tween 20 in PBS), and then blocked with 4% bovine serum albumin in PBS (supplemented with 0.05% Tween 20) for 2 h at room temperature. The plate was again washed five times, before incubating for 90 min at room temperature with 10% serum in the aforementioned blocking buffer (without Tween 20). The plate was washed five times, and then incubated for 90 min at room temperature with 10 × anti-DNA antibody (HRP-conjugated; from the Cell Death kit) diluted 1:100 in blocking buffer. After five more washes, the plate was developed with 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (Invitrogen) followed by a 2 N sulfuric acid stop solution. Absorbance was measured at a wavelength of 450 nm using a Cytation 5 Cell Imaging Multi-Mode Reader (BioTek). Data were normalized to in vitro-prepared NET standards included on every plate.
Quantification of citrullinated-histone H3
Citrullinated-histone H3 was quantified in sera using the Citrullinated Histone H3 (Clone 11D3) ELISA Kit (Cayman, 501620) according to the manufacturer’s instructions.
Statistical analysis
When two groups were present, normally-distributed data were analyzed by two-sided t test and skewed data were analyzed by Mann–Whitney test. Correlations were tested by Pearson’s correlation coefficient in normally-distributed data and Spearman’s correlation coefficient in skewed data. Data analysis was with GraphPad Prism software version 8. Statistical significance was defined as p < 0.05.
Results
We identified 11 patients who developed a thrombotic event while admitted for treatment of COVID-19 at a large academic medical center (Table 1). The events consisted of three strokes and nine VTE events (Table 2). Nine (82%) were receiving at least prophylactic doses of heparinoids at the time the event was diagnosed. The other two patients were found to have pulmonary emboli on the day of admission (Table 2). Nine of the patients were receiving respiratory support by mechanical ventilation at the time of the event. At the time of this analysis, eight patients had been discharged from the hospital, two had died, and one remained hospitalized (Table 2).
To better understand the extent to which patients diagnosed with thrombotic events differed from other patients hospitalized with COVID-19, we identified a matched cohort of 33 patients hospitalized with COVID-19 over the same month (Table 1). The control cohort was identical for age and sex. The cohort was similar in terms of comorbidities and ultimate outcome (18% died in the thrombosis group and 21% in the control group). Thirty of those matched COVID-19 control patients (91%) were receiving at least prophylactic doses of heparinoids. While no significant difference was appreciated in oxygenation efficiency (SpO2/FiO2) between the groups at the time of sample collection, a higher proportion of thrombosis patients required mechanical ventilation during hospitalization (Table 1). For all 44 patients, we were able to access a blood sample collected during their hospitalization. As it relates to diagnosis of the first thrombotic event for each patient, six blood samples were banked within three days of event diagnosis. Three were banked earlier (5, 7, and 12 days prior to event diagnosis) and two later (both at 6 days). For 33 matched control blood samples, ten were banked on admission, ten were banked on hospital day 1, four on hospital day 2, and one on hospital day 3. The remaining were banked between hospital days 4 and 12. As compared with the control group, patients with a thrombotic event demonstrated significantly higher levels of calprotectin, a marker of neutrophil activation (Fig. 1a). Similarly, three different markers of NETs (cell-free DNA, myeloperoxidase-DNA complexes, and citrullinated histone H3) were also markedly elevated in the thrombosis group as compared with the matched controls (Fig. 1b–d).
Elevated levels of NETs in the blood of COVID-19 patients diagnosed with a thrombotic event, as compared with matched controls. Serum was tested for calprotectin (a), cell-free DNA (b), myeloperoxidase-DNA complexes (c), and citrullinated histone H3 (d). N = 33 for the control group and n = 11 for the thrombosis group. Comparisons were by Mann–Whitney test; *p < 0.05, **p < 0.01, and ***p < 0.001
We then turned our attention to other clinical biomarkers that might associate with a thrombotic event. The thrombosis group had higher levels of peak D-dimer (Fig. 2a), but not troponin (Fig. 2b). Peak CRP and ferritin were also modestly higher in the thrombosis group (Fig. 2c, d). Interestingly, peak neutrophil levels did not differ between groups, but platelets were significantly higher in patients who developed thrombosis (Fig. 2e, f). Finally, we asked whether there was an association between blood markers of neutrophil activation (such as calprotectin and cell-free DNA) and D-dimer within this cohort of COVID-19 patients (n = 44). Despite the small number of patients, there were strong correlations between peak D-dimer and calprotectin, cell-free DNA (Fig. 3a, b), and peak neutrophil count (Supplementary Figure S1). Levels of calprotectin and cell-free DNA also correlated positively with peak platelet counts (Supplementary Figure S2). Calprotectin and cell-free DNA also predicted worse oxygen efficiency in this cohort (Supplementary Figure S3).
Association between peak levels of clinical biomarkers and diagnosis of a thrombotic event. Clinical testing is reported for D-dimer (a), troponin (b), C-reactive protein (c), ferritin (d), absolute neutrophil count (e), and absolute platelet count (f). N = 33 for the control group and n = 11 for the thrombosis group. Comparisons were by Mann–Whitney test; *p < 0.05 and **p < 0.01. Comparisons for peak troponin and peak neutrophil count were not statistically significant
Discussion
Hyperactivity of the coagulation system is a common finding of severe COVID-19 [29]. Indeed, many patients have a profile to suggest a prothrombotic diathesis including high levels of fibrin degradation products (D-dimer), elevated fibrinogen levels, and low antithrombin levels [29, 30]. We recently characterized the blood of 50 patients hospitalized with COVID-19 and found significantly elevated levels of NETs as compared with healthy controls [19]. Here, we extend that analysis by demonstrating particularly high levels of NETs in a subgroup of patients with COVID-19 who were clinically diagnosed with at least one thrombotic event. This study is limited by its small sample size and inherent effect of utilizing discarded specimens at various sampling time points. Given the known link between NETs and thrombosis in many inflammatory conditions, these data suggest that the role of NETs in COVID-19-associated thrombophilia warrants systematic and prospective investigation.
Intravascular NET release is responsible for initiation and accretion of thrombotic events in arteries, veins, and microvessels, where thrombotic disease can drive end-organ damage in lungs, heart, kidneys, and other organs [31, 32]. Mechanistically, DNA in NETs may directly activate the extrinsic pathway of coagulation [33], while NETs also present tissue factor to initiate the intrinsic pathway [30,31,35]. Serine proteases in NETs such as neutrophil elastase promote coagulation by proteolyzing various tissue factor pathway inhibitors [36]. There are likely synergistic mechanisms by which SARS-CoV-2 infection may result in macrovascular and microvascular occlusions. These include autoantibody- and cytokine-mediated activation of innate immune cells including neutrophils and platelets, vasoconstriction in the setting of hypoxia, and direct activation of endothelial cells by viral infection [21, 25, 28, 37, 38]. Here, we not only demonstrate markedly elevated NETs among patients with COVID-19 and clinically diagnosed thrombosis, but also a strong correlation between neutrophil-activation markers/NETs with D-dimer, a degradation product of fibrin. These data suggest a potential clinically actionable link between NET formation and enhanced coagulation turnover that may contribute to the COVID-19 prothrombotic state. We speculate that tripartite interplay between neutrophils, endothelial cells, and platelets might be critical for COVID-19-associated thrombosis as has been characterized in models of other thrombo-inflammatory diseases [32, 33].
Approaches to combatting NETs [22, 39, 40] include the dismantling of NETs with deoxyribonucleases and strategies that prevent initiation of NET release such as neutrophil elastase inhibitors, peptidylarginine deiminase 4 inhibitors, and adenosine receptor agonists such as dipyridamole (eg. NCT04391179) [41]. As we await definitive antiviral solutions to the current pandemic, we posit that anti-neutrophil therapies may be part of a personalized strategy for some individuals affected by COVID-19. Furthermore, those patients with hyperactive neutrophils may be at particularly high risk for thrombotic events and might therefore benefit from more aggressive anticoagulation while hospitalized.
References
Pedersen SF, Ho YC (2020) SARS-CoV-2: a storm is raging. J Clin Invest. https://doi.org/10.1172/JCI137647
Zhang B, Zhou X, Zhu C, Feng F, Qiu Y, Feng J, Jia Q, Song Q, Zhu B, Wang J (2020) Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19.2020.2003.2012.20035048. doi:https://doi.org/10.1101/2020.03.12.20035048 %J medRxiv
Song C-Y, Xu J, He J-Q, Lu Y-Q (2020) COVID-19 early warning score: a multi-parameter screening tool to identify highly suspected patients.2020.2003.2005.20031906. doi:https://doi.org/10.1101/2020.03.05.20031906 %J medRxiv
Iba T, Levy JH, Raj A, Warkentin TE (2019) Advance in the management of sepsis-induced coagulopathy and disseminated intravascular coagulation. J Clin Med. https://doi.org/10.3390/jcm8050728
Ward PA, Fattahi F (2019) New strategies for treatment of infectious sepsis. J Leukoc Biol 106(1):187–192. https://doi.org/10.1002/JLB.4MIR1118-425R
Ali RA, Gandhi AA, Meng H, Yalavarthi S, Vreede AP, Estes SK, Palmer OR, Bockenstedt PL, Pinsky DJ, Greve JM, Diaz JA, Kanthi Y, Knight JS (2019) Adenosine receptor agonism protects against NETosis and thrombosis in antiphospholipid syndrome. Nat Commun 10(1):1916. https://doi.org/10.1038/s41467-019-09801-x
Meng H, Yalavarthi S, Kanthi Y, Mazza LF, Elfline MA, Luke CE, Pinsky DJ, Henke PK, Knight JS (2017) In vivo role of neutrophil extracellular traps in antiphospholipid antibody-mediated venous thrombosis. Arthritis Rheumatol 69(3):655–667. https://doi.org/10.1002/art.39938
Yadav V, Chi L, Zhao R, Tourdot BE, Yalavarthi S, Jacobs BN, Banka A, Liao H, Koonse S, Anyanwu AC, Visovatti SH, Holinstat MA, Kahlenberg JM, Knight JS, Pinsky DJ, Kanthi Y (2019) Ectonucleotidase tri(di)phosphohydrolase-1 (ENTPD-1) disrupts inflammasome/interleukin 1beta-driven venous thrombosis. J Clin Invest 129(7):2872–2877. https://doi.org/10.1172/JCI124804
Potey PM, Rossi AG, Lucas CD, Dorward DA (2019) Neutrophils in the initiation and resolution of acute pulmonary inflammation: understanding biological function and therapeutic potential. J Pathol 247(5):672–685. https://doi.org/10.1002/path.5221
Frantzeskaki F, Armaganidis A, Orfanos SE (2017) Immunothrombosis in acute respiratory distress syndrome: cross talks between inflammation and coagulation. Respiration 93(3):212–225. https://doi.org/10.1159/000453002
Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A (2004) Neutrophil extracellular traps kill bacteria. Science 303(5663):1532–1535. https://doi.org/10.1126/science.1092385
Warnatsch A, Ioannou M, Wang Q, Papayannopoulos V (2015) Inflammation: Neutrophil extracellular traps license macrophages for cytokine production in atherosclerosis. Science 349(6245):316–320. https://doi.org/10.1126/science.aaa8064
Doring Y, Soehnlein O, Weber C (2017) Neutrophil extracellular traps in atherosclerosis and atherothrombosis. Circ Res 120(4):736–743. https://doi.org/10.1161/CIRCRESAHA.116.309692
Brill A, Fuchs TA, Savchenko AS, Thomas GM, Martinod K, De Meyer SF, Bhandari AA, Wagner DD (2012) Neutrophil extracellular traps promote deep vein thrombosis in mice. J Thromb Haemost 10(1):136–144. https://doi.org/10.1111/j.1538-7836.2011.04544.x
Tanaka K, Koike Y, Shimura T, Okigami M, Ide S, Toiyama Y, Okugawa Y, Inoue Y, Araki T, Uchida K, Mohri Y, Mizoguchi A, Kusunoki M (2014) In vivo characterization of neutrophil extracellular traps in various organs of a murine sepsis model. PLoS ONE 9(11):e111888. https://doi.org/10.1371/journal.pone.0111888
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395(10229):1054–1062. https://doi.org/10.1016/S0140-6736(20)30566-3
Bunce PE, High SM, Nadjafi M, Stanley K, Liles WC, Christian MD (2011) Pandemic H1N1 influenza infection and vascular thrombosis. Clin Infect Dis 52(2):e14-17. https://doi.org/10.1093/cid/ciq125
Colling ME, Kanthi Y (2020) COVID–19-associated coagulopathy: An exploration of mechanisms. Vascular Medicine 25(5):471–478
Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA, Blair CN, Weber A, Barnes BJ, Egeblad M, Woods RJ, Kanthi Y, Knight JS (2020) Neutrophil extracellular traps in COVID-19. JCI Insight. https://doi.org/10.1172/jci.insight.138999
Middleton EA, He XY, Denorme F, Campbell RA, Ng D, Salvatore SP, Mostyka M, Baxter-Stoltzfus A, Borczuk AC, Loda M, Cody MJ, Manne BK, Portier I, Harris ES, Petrey AC, Beswick EJ, Caulin AF, Iovino A, Abegglen LM, Weyrich AS, Rondina MT, Egeblad M, Schiffman JD, Yost CC (2020) Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood 136(10):1169–1179. https://doi.org/10.1182/blood.2020007008
Leppkes M, Knopf J, Naschberger E, Lindemann A, Singh J, Herrmann I, Sturzl M, Staats L, Mahajan A, Schauer C, Kremer AN, Volkl S, Amann K, Evert K, Falkeis C, Wehrfritz A, Rieker RJ, Hartmann A, Kremer AE, Neurath MF, Munoz LE, Schett G, Herrmann M (2020) Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine 58:102925. https://doi.org/10.1016/j.ebiom.2020.102925
Barnes BJ, Adrover JM, Baxter-Stoltzfus A, Borczuk A, Cools-Lartigue J, Crawford JM, Dassler-Plenker J, Guerci P, Huynh C, Knight JS, Loda M, Looney MR, McAllister F, Rayes R, Renaud S, Rousseau S, Salvatore S, Schwartz RE, Spicer JD, Yost CC, Weber A, Zuo Y, Egeblad M (2020) Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med. https://doi.org/10.1084/jem.20200652
Skendros P, Mitsios A, Chrysanthopoulou A, Mastellos DC, Metallidis S, Rafailidis P, Ntinopoulou M, Sertaridou E, Tsironidou V, Tsigalou C, Tektonidou MG, Konstantinidis T, Papagoras C, Mitroulis I, Germanidis G, Lambris JD, Ritis K (2020) Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J Clin Invest. https://doi.org/10.1172/JCI141374
Radermecker C, Detrembleur N, Guiot J, Cavalier E, Henket M, d’Emal C, Vanwinge C, Cataldo D, Oury C, Delvenne P, Marichal T (2020) Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J Exp Med. https://doi.org/10.1084/jem.20201012
Nicolai L, Leunig A, Brambs S, Kaiser R, Weinberger T, Weigand M, Muenchhoff M, Hellmuth JC, Ledderose S, Schulz H, Scherer C, Rudelius M, Zoller M, Hochter D, Keppler O, Teupser D, Zwissler B, von Bergwelt-Baildon M, Kaab S, Massberg S, Pekayvaz K, Stark K (2020) Immunothrombotic dysregulation in COVID-19 pneumonia is associated with respiratory failure and coagulopathy. Circulation 142(12):1176–1189. https://doi.org/10.1161/CIRCULATIONAHA.120.048488
Shi H, Zuo Y, Yalavarthi S, Gockman K, Zuo M, Madison JA, Blair C, Woodward W, Lezak SP, Lugogo NL, Woods RJ, Lood C, Knight JS, Kanthi Y (2020) Neutrophil calprotectin identifies severe pulmonary disease in COVID-19. J Leukoc Biol. https://doi.org/10.1002/JLB.3COVCRA0720-359R
Kessenbrock K, Krumbholz M, Schonermarck U, Back W, Gross WL, Werb Z, Grone HJ, Brinkmann V, Jenne DE (2009) Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med 15(6):623–625. https://doi.org/10.1038/nm.1959nm.1959[pii]
Zuo Y, Estes SK, Ali RA, Gandhi AA, Yalavarthi S, Shi H, Sule G, Gockman K, Madison JA, Zuo M, Yadav V, Wang J, Woodard W, Lezak SP, Lugogo NL, Smith SA, Morrissey JH, Kanthi Y, Knight JS (2020) Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci Transl Med:eabd3876. https://doi.org/10.1126/scitranslmed.abd3876
Tang N, Li D, Wang X, Sun Z (2020) Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost 18(4):844–847. https://doi.org/10.1111/jth.14768
Han H, Yang L, Liu R, Liu F, Wu KL, Li J, Liu XH, Zhu CL (2020) Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clin Chem Lab Med. https://doi.org/10.1515/cclm-2020-0188
Pfeiler S, Massberg S, Engelmann B (2014) Biological basis and pathological relevance of microvascular thrombosis. Thromb Res 133(Suppl 1):S35-37. https://doi.org/10.1016/j.thromres.2014.03.016
Ma AC, Kubes P (2008) Platelets, neutrophils, and neutrophil extracellular traps (NETs) in sepsis. J Thromb Haemost 6(3):415–420. https://doi.org/10.1111/j.1538-7836.2007.02865.x
Gould TJ, Vu TT, Swystun LL, Dwivedi DJ, Mai SH, Weitz JI, Liaw PC (2014) Neutrophil extracellular traps promote thrombin generation through platelet-dependent and platelet-independent mechanisms. Arterioscler Thromb Vasc Biol 34(9):1977–1984. https://doi.org/10.1161/ATVBAHA.114.304114
Kambas K, Chrysanthopoulou A, Vassilopoulos D, Apostolidou E, Skendros P, Girod A, Arelaki S, Froudarakis M, Nakopoulou L, Giatromanolaki A, Sidiropoulos P, Koffa M, Boumpas DT, Ritis K, Mitroulis I (2014) Tissue factor expression in neutrophil extracellular traps and neutrophil derived microparticles in antineutrophil cytoplasmic antibody associated vasculitis may promote thromboinflammation and the thrombophilic state associated with the disease. Ann Rheum Dis 73(10):1854–1863. https://doi.org/10.1136/annrheumdis-2013-203430
Wang Y, Luo L, Braun OO, Westman J, Madhi R, Herwald H, Morgelin M, Thorlacius H (2018) Neutrophil extracellular trap-microparticle complexes enhance thrombin generation via the intrinsic pathway of coagulation in mice. Sci Rep 8(1):4020. https://doi.org/10.1038/s41598-018-22156-5
Massberg S, Grahl L, von Bruehl ML, Manukyan D, Pfeiler S, Goosmann C, Brinkmann V, Lorenz M, Bidzhekov K, Khandagale AB, Konrad I, Kennerknecht E, Reges K, Holdenrieder S, Braun S, Reinhardt C, Spannagl M, Preissner KT, Engelmann B (2010) Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat Med 16(8):887–896. https://doi.org/10.1038/nm.2184
Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka F, Moch H (2020) Endothelial cell infection and endotheliitis in COVID-19. Lancet 395(10234):1417–1418. https://doi.org/10.1016/S0140-6736(20)30937-5
Nakazawa D, Ishizu A (2020) Immunothrombosis in severe COVID-19. EBioMedicine 59:102942. https://doi.org/10.1016/j.ebiom.2020.102942
Thalin C, Hisada Y, Lundstrom S, Mackman N, Wallen H (2019) Neutrophil extracellular traps: villains and targets in arterial, venous, and cancer-associated thrombosis. Arterioscler Thromb Vasc Biol 39(9):1724–1738. https://doi.org/10.1161/ATVBAHA.119.312463
Barnado A, Crofford LJ, Oates JC (2016) At the Bedside: Neutrophil extracellular traps (NETs) as targets for biomarkers and therapies in autoimmune diseases. J Leukoc Biol 99(2):265–278. https://doi.org/10.1189/jlb.5BT0615-234R
Kanthi Y, Knight JS, Zuo Y, Pinsky DJ (2020) New (re)purpose for an old drug: purinergic modulation may extinguish the COVID-19 thromboinflammatory firestorm. JCI Insight 5(14)
Acknowledgements
The work was supported by a COVID-19 Cardiovascular Impact Research Ignitor Grant from the Michigan Medicine Frankel Cardiovascular Center as well as by the A. Alfred Taubman Medical Research Institute. YZ was supported by career development grants from the Rheumatology Research Foundation. JAM was partially supported by the VA Healthcare System. JSK was supported by grants from the NIH (R01HL115138), Lupus Research Alliance, and Burroughs Wellcome Fund. YK was supported by the Intramural Research Program of the NIH and National Heart, Lung, and Blood Institute, the Falk Medical Research Trust Catalyst Award, JOBST-American Venous Forum Award, and NIH (K08HL131993, R01HL150392).
Author information
Authors and Affiliations
Contributions
YZ, MZ, SY, KG, JM, HS, WW, and SPL conducted experiments and analyzed data. YZ, MZ, NLL, YK, and JSK conceived the study and analyzed data. All authors participated in writing the manuscript and gave approval before submission.
Corresponding authors
Ethics declarations
Conflict of interest
YK has received consulting fees from Surface Oncology, and has a pending patent on the use of biogases in vascular disease. JSK has received grant support from Jazz Pharmaceuticals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zuo, Y., Zuo, M., Yalavarthi, S. et al. Neutrophil extracellular traps and thrombosis in COVID-19. J Thromb Thrombolysis 51, 446–453 (2021). https://doi.org/10.1007/s11239-020-02324-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11239-020-02324-z