Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs
Abstract
:1. Introduction
2. Materials and Methods
2.1. SARS-CoV-2 Genomic Analysis
2.2. MiRNA Binding Site Prediction
2.3. MiRNA Expression in Human Tissue and Cell Lines
2.4. Statistical Analysis
3. Results
3.1. Globally Representative SARS-CoV-2 Genomes Demonstrate Minimal Variation in Nucleotide Sequence and Predicted miRNA Binding Sites
3.2. Genomic Regions of SARS-CoV-2 Demonstrate Unique Predicted miRNA Binding Profiles
3.3. Expression of miRNAs in Lungs Predicted to Bind SARS-CoV-2
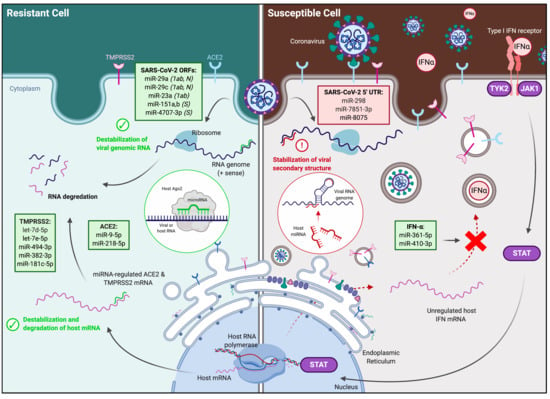
3.4. Identification and Expression of Predicted miRNAs in SARS-CoV-2-Resistant and -Susceptible Cells
3.5. Identification of Predicted miRNAs to ACE2 and TMPRSS2 in SARS-CoV-2-Resistant and -Susceptible Cells
3.6. Identification of Predicted miRNAs to IFN Genes in SARS-CoV-2-Resistant and -Susceptible Cells
4. Discussion
4.1. MiRNAs Targeting the SARS-CoV-2 Viral Genome
4.2. MiRNAs Targeting Viral Entry Proteins and IFNs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 1 May 2020).
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-l.; Hui, D.S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, C.; Allon, S.J.; Nyquist, S.K.; Mbano, I.; Miao, V.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; Feldman, J.; et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is enriched in specific cell subsets across tissues. Cell 2020, 181, 1016–1035.e19. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Sampaziotis, F. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef] [Green Version]
- Lukassen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Meister, M.; Veith, C.; Boots, A.W. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020, 39, e105114. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Chan, J.; Yuen, T.; Shuai, H.; Yuan, S.; Wang, Y.; Hu, B.; Yip, C.; Tsang, J.; Huang, X. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study. Lancet Microbe 2020, 1, E14–E23. [Google Scholar] [CrossRef]
- Gebert, L.F.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Kincaid, R.P.; Sullivan, C.S. Virus-encoded microRNAs: An overview and a look to the future. PLoS Pathog. 2012, 8, e1003018. [Google Scholar] [CrossRef] [Green Version]
- Dölken, L.; Krmpotic, A.; Kothe, S.; Tuddenham, L.; Tanguy, M.; Marcinowski, L.; Ruzsics, Z.; Elefant, N.; Altuvia, Y.; Margalit, H. Cytomegalovirus microRNAs facilitate persistent virus infection in salivary glands. PLoS Pathog. 2010, 6, e1001150. [Google Scholar] [CrossRef] [Green Version]
- Bauman, Y.; Nachmani, D.; Vitenshtein, A.; Tsukerman, P.; Drayman, N.; Stern-Ginossar, N.; Lankry, D.; Gruda, R.; Mandelboim, O. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe 2011, 9, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017, 23, 80–93. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Hatcher, E.L.; Zhdanov, S.A.; Bao, Y.; Blinkova, O.; Nawrocki, E.P.; Ostapchuck, Y.; Schäffer, A.A.; Brister, J.R. Virus Variation Resource–improved response to emergent viral outbreaks. Nucleic Acids Res. 2017, 45, D482–D490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, Q.; Zhang, Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020, 30, 1346–1351. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Nagilla, P.; Le, H.-S.; Bunney, C.; Zych, C.; Thalamuthu, A.; Bar-Joseph, Z.; Mathavan, S.; Ayyavoo, V. Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1). PLoS ONE 2011, 6, e22730. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Zuker, M.; Stiegler, P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981, 9, 133–148. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with 2019 Novel Coronavirus Disease, United States. Emerg. Infect. Dis. 2020, 26. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schäfer, A.; Dinnon, K.H.; Stevens, L.J. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, 12, eabb5883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyler, E.; Mösbauer, K.; Franke, V.; Diag, A.; Gottula, L.T.; Arsie, R.; Klironomos, F.; Koppstein, D.; Ayoub, S.; Buccitelli, C.; et al. Bulk and single-cell gene expression profiling of SARS-CoV-2 infected human cell lines identifies molecular targets for therapeutic intervention. bioRxiv 2020. [Google Scholar] [CrossRef]
- Müller, S.; Glaß, M.; Singh, A.K.; Haase, J.; Bley, N.; Fuchs, T.; Lederer, M.; Dahl, A.; Huang, H.; Chen, J. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A-and miRNA-dependent manner. Nucleic Acids Res. 2019, 47, 375–390. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Chu, H.; Wen, L.; Shuai, H.; Yang, D.; Wang, Y.; Hou, Y.; Zhu, Z.; Yuan, S.; Yin, F. Competing endogenous RNA network profiling reveals novel host dependency factors required for MERS-CoV propagation. Emerg. Microbes Infect. 2020, 9, 733–746. [Google Scholar] [CrossRef]
- Wei, P.; Xie, Y.; Abel, P.W.; Huang, Y.; Ma, Q.; Li, L.; Hao, J.; Wolff, D.W.; Wei, T.; Tu, Y. Transforming growth factor (TGF)-β1-induced miR-133a inhibits myofibroblast differentiation and pulmonary fibrosis. Cell Death Dis. 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Fukuhara, T.; Kambara, H.; Shiokawa, M.; Ono, C.; Katoh, H.; Morita, E.; Okuzaki, D.; Maehara, Y.; Koike, K.; Matsuura, Y. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with hepatitis C virus. J. Virol. 2012, 86, 7918–7933. [Google Scholar] [CrossRef] [Green Version]
- Schult, P.; Roth, H.; Adams, R.L.; Mas, C.; Imbert, L.; Orlik, C.; Ruggieri, A.; Pyle, A.M.; Lohmann, V. microRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Wen, W.; He, Z.; Jing, Q.; Hu, Y.; Lin, C.; Zhou, R.; Wang, X.; Su, Y.; Yuan, J.; Chen, Z. Cellular microRNA-miR-548g-3p modulates the replication of dengue virus. J. Infect. 2015, 70, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Trobaugh, D.W.; Gardner, C.L.; Sun, C.; Haddow, A.D.; Wang, E.; Chapnik, E.; Mildner, A.; Weaver, S.C.; Ryman, K.D.; Klimstra, W.B. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 2014, 506, 245–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.; Leibowitz, J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015, 206, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef] [PubMed]
- Lambert, D.W.; Yarski, M.; Warner, F.J.; Thornhill, P.; Parkin, E.T.; Smith, A.I.; Hooper, N.M.; Turner, A.J. Tumor necrosis factor-α convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J. Biol. Chem. 2005, 280, 30113–30119. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.P.; Look, D.C.; Shi, L.; Hickey, M.; Pewe, L.; Netland, J.; Farzan, M.; Wohlford-Lenane, C.; Perlman, S.; McCray, P.B. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005, 79, 14614–14621. [Google Scholar] [CrossRef] [Green Version]
- Tseng, C.-T.K.; Tseng, J.; Perrone, L.; Worthy, M.; Popov, V.; Peters, C.J. Apical entry and release of severe acute respiratory syndrome-associated coronavirus in polarized Calu-3 lung epithelial cells. J. Virol. 2005, 79, 9470–9479. [Google Scholar] [CrossRef] [Green Version]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhu, L.; Liao, S.; Xu, Z.; Zhou, Y. The porcine microRNA transcriptome response to transmissible gastroenteritis virus infection. PLoS ONE 2015, 10, e0120377. [Google Scholar] [CrossRef] [Green Version]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanford, R.E.; Hildebrandt-Eriksen, E.S.; Petri, A.; Persson, R.; Lindow, M.; Munk, M.E.; Kauppinen, S.; Ørum, H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 2010, 327, 198–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Wang, F.; Argyris, E.; Chen, K.; Liang, Z.; Tian, H.; Huang, W.; Squires, K.; Verlinghieri, G.; Zhang, H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007, 13, 1241–1247. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.-P.; Dai, H.-J.; Yang, Y.-H.; Zhuang, Y.; Sheng, R. MicroRNA-23b inhibits enterovirus 71 replication through downregulation of EV71 VPl protein. Intervirology 2013, 56, 195–200. [Google Scholar] [CrossRef]
- Bai, X.T.; Nicot, C. miR-28-3p is a cellular restriction factor that inhibits human T cell leukemia virus, type 1 (HTLV-1) replication and virus infection. J. Biol. Chem. 2015, 290, 5381–5390. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.-L.; Li, Y.-X.; Zheng, S.-Q.; Liu, M.; Li, X.; Tang, H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antivir. Res. 2010, 88, 169–175. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, Q.; Zhang, T. Pangolin homology associated with 2019-nCoV. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Nao, N.; Yamagishi, J.; Miyamoto, H.; Igarashi, M.; Manzoor, R.; Ohnuma, A.; Tsuda, Y.; Furuyama, W.; Shigeno, A.; Kajihara, M. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. MBio 2017, 8, e02298-16. [Google Scholar] [CrossRef] [Green Version]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020, 584, 154–156. [Google Scholar] [CrossRef]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef]
- Sun, J.-Z.; Wang, J.; Wang, S.; Yuan, D.; Li, Z.; Yi, B.; Hou, Q.; Mao, Y.; Liu, W. MicroRNA miR-320a and miR-140 inhibit mink enteritis virus infection by repression of its receptor, feline transferrin receptor. Virol. J. 2014, 11, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duy, J.; Koehler, J.W.; Honko, A.N.; Schoepp, R.J.; Wauquier, N.; Gonzalez, J.-P.; Pitt, M.L.; Mucker, E.M.; Johnson, J.C.; O’Hearn, A. Circulating microRNA profiles of Ebola virus infection. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Holland, J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-S.; Gong, J.-N.; Yu, J.; Wang, F.; Zhang, X.-H.; Yin, X.-L.; Tan, Z.-Q.; Luo, Z.-M.; Yang, G.-H.; Shen, C. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood 2012, 119, 4992–5004. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Li, X.; He, Q.; Gao, J.; Gao, Y.; Liu, B.; Liu, F. miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res. 2013, 23, 1356–1368. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Wang, X.-S.; Yang, G.-H.; Zhai, P.-F.; Xiao, Z.; Xia, L.-Y.; Chen, L.-R.; Wang, Y.; Wang, X.-Z.; Bi, L.-X. miR-29a and miR-142-3p downregulation and diagnostic implication in human acute myeloid leukemia. Mol. Biol. Rep. 2012, 39, 2713–2722. [Google Scholar] [CrossRef]
- Xu, L.; Xu, Y.; Jing, Z.; Wang, X.; Zha, X.; Zeng, C.; Chen, S.; Yang, L.; Luo, G.; Li, B. Altered expression pattern of miR-29a, miR-29b and the target genes in myeloid leukemia. Exp. Hematol. Oncol. 2014, 3, 17. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-Y.; Lee, J.H.; Ha, M.; Nam, J.-W.; Kim, V.N. miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat. Struct. Mol. Biol. 2009, 16, 23. [Google Scholar] [CrossRef]
- Gong, J.; Yu, J.; Lin, H.; Zhang, X.; Yin, X.; Xiao, Z.; Wang, F.; Wang, X.; Su, R.; Shen, C. The role, mechanism and potentially therapeutic application of microRNA-29 family in acute myeloid leukemia. Cell Death Differ. 2014, 21, 100–112. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Fang, J.H.; Yun, J.P.; Yang, J.; Zhang, Y.; Jia, W.H.; Zhuang, S.M. Effects of MicroRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 2010, 51, 836–845. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Croce, C.M. Role of miR-15/16 in CLL. Cell Death Differ. 2015, 22, 6–11. [Google Scholar] [CrossRef] [Green Version]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ofir, M.; Hacohen, D.; Ginsberg, D. MiR-15 and miR-16 are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol. Cancer Res. 2011, 9, 440–447. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Zhang, Z.; Yang, X.; Yang, W.; Xue, C.; Cao, X. miR-23b suppresses lung carcinoma cell proliferation through CCNG1. Oncol. Lett. 2018, 16, 4317–4324. [Google Scholar] [CrossRef]
- Luna, J.M.; Scheel, T.K.; Danino, T.; Shaw, K.S.; Mele, A.; Fak, J.J.; Nishiuchi, E.; Takacs, C.N.; Catanese, M.T.; de Jong, Y.P. Hepatitis C virus RNA functionally sequesters miR-122. Cell 2015, 160, 1099–1110. [Google Scholar] [CrossRef] [Green Version]
- Fulzele, S.; Sahay, B.; Yusufu, I.; Lee, T.J.; Sharma, A.; Kolhe, R.; Isales, C.M. COVID-19 Virulence in Aged Patients Might Be Impacted by the Host Cellular MicroRNAs Abundance/Profile. Aging Dis. 2020, 11, 509. [Google Scholar] [CrossRef]
- Chow, J.T.-S.; Salmena, L. Prediction and Analysis of SARS-CoV-2-Targeting MicroRNA in Human Lung Epithelium. Genes 2020, 11, 1002. [Google Scholar] [CrossRef]
- Cullen, B.R. Viruses and microRNAs. Nat. Genet. 2006, 38, S25–S30. [Google Scholar] [CrossRef]
- Demirci, M.D.S.; Adan, A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ 2020, 8, e9369. [Google Scholar] [CrossRef] [PubMed]
- Morales, L.; Oliveros, J.C.; Fernandez-Delgado, R.; Robert tenOever, B.; Enjuanes, L.; Sola, I. SARS-CoV-encoded small RNAs contribute to infection-associated lung pathology. Cell Host Microbe 2017, 21, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Arisan, E.D.; Dart, A.; Grant, G.H.; Arisan, S.; Cuhadaroglu, S.; Lange, S.; Uysal-Onganer, P. The Prediction of miRNAs in SARS-CoV-2 Genomes: Hsa-miR Databases Identify 7 Key miRs Linked to Host Responses and Virus Pathogenicity-Related KEGG Pathways Significant for Comorbidities. Viruses 2020, 12, 614. [Google Scholar] [CrossRef] [PubMed]
- Periwal, N.; Sarma, S.; Arora, P.; Sood, V. In-silico analysis of SARS-CoV-2 genomes: Insights from SARS encoded non-coding RNAs. bioRxiv 2020. [Google Scholar] [CrossRef]
- Choi, E.-J.; Kim, H.B.; Baek, Y.H.; Kim, E.-H.; Pascua, P.N.Q.; Park, S.-J.; Kwon, H.-i.; Lim, G.-J.; Kim, S.; Kim, Y.-I. Differential microRNA expression following infection with a mouse-adapted, highly virulent avian H5N2 virus. BMC Microbiol. 2014, 14, 252. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Ouyang, X.; Jiang, X.; Gu, D.; Lin, Y.; Kong, S.; Xie, W. Dysregulated serum microRNA expression profile and potential biomarkers in hepatitis C virus-infected patients. Int. J. Med Sci. 2015, 12, 590. [Google Scholar] [CrossRef] [Green Version]
- Ling, T.-Y.; Kuo, M.-D.; Li, C.-L.; Alice, L.Y.; Huang, Y.-H.; Wu, T.-J.; Lin, Y.-C.; Chen, S.-H.; Yu, J. Identification of pulmonary Oct-4+ stem/progenitor cells and demonstration of their susceptibility to SARS coronavirus (SARS-CoV) infection in vitro. Proc. Natl. Acad. Sci. USA 2006, 103, 9530–9535. [Google Scholar] [CrossRef] [Green Version]
- Mallick, B.; Ghosh, Z.; Chakrabarti, J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS ONE 2009, 4, e7837. [Google Scholar] [CrossRef]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef]








| Location | Wuhan, China (RefSeq) | Washington, USA | Washington, USA | California, USA | Washington, USA | Punjab, Pakistan |
|---|---|---|---|---|---|---|
| GenBank | MN908947.3 | MT246485.1 | MT345855.1 | MT258382.1 | MT293170.1 | MT262993.1 |
| Overall | ||||||
| Bases | 29,903 | 29,775 | 29,859 | 29,885 | 29,798 | 29,836 |
| Sequence identity a | - | 99.85% b | 99.85% b | 99.87% b | 99.88% b | 99.88% |
| 5′ UTR | ||||||
| Bases | 265 | 137 | 234 | 247 | 151 | 265 |
| Sequence identity a | - | 82.5% b | 91.45% b | 99.60 | 92.72% b | 100% |
| Mutations | - | - | C→T | C→T | - | - |
| S protein | ||||||
| Bases | 3822 | 3822 | 3822 | 3822 | 3822 | 3822 |
| Sequence identity a | - | 100% | 100% | 99.95% | 100% | 100% |
| Mutations | - | - | - | T→W (either T or A); A→G | - | - |
| 3′ UTR | ||||||
| Bases | 229 | 229 | 198 | 229 | 238 | 198 |
| Sequence identity a | - | 98.69% | 100% | 93.45% b | 97.38% b | 100% |
| Mutations | - | TGAC → AAAA | - | G→R (either G or A); C→M (either C or A) | 9 nt insertion in poly-A tail | - |
| Source | ACE2/TMPRSS2 Expression | SARS-CoV-2 Infectivity | |
|---|---|---|---|
| Huh7 [8,14,32,33] | Liver | High [47] | Moderate |
| A549 [8,14,32,33] | Lung | Variable [48] | Low to Moderate |
| Calu-3 [8,14,32,34] | Lung | High [49] | High |
| Primary human lung fibroblasts [12,13] | Lung | None | Low |
| miRNA | Log2-Fold Change | p-value | Favors a |
|---|---|---|---|
| ORF1ab | |||
| miR-139-5p | 0.945 | 0.0305 | Resistant |
| miR-664b-3p | 0.733 | 0.0206 | - |
| miR-23a-3p | −0.761 | 0.0015 | Resistant |
| miR-548d-3p | −0.807 | 0.0517 | - |
| miR-15b-5p | −0.812 | 0.0001 | - |
| miR-23c | −0.910 | 0.0049 | - |
| miR-23b-3p | −0.991 | <0.0001 | Resistant |
| miR-374b-3p | −1.069 | 0.0217 | - |
| miR-374a-3p | −1.582 | 0.0001 | - |
| Nucleocapsid | |||
| miR-103a-3p | 0.904 | 0.0005 | - |
| miR-107 | 1.040 | 0.0003 | - |
| ACE2 | |||
| miR-483-3p | 4.460 | <0.0001 | Susceptible |
| miR-4463 | 3.048 | <0.0001 | - |
| TMPRSS2 | |||
| miR-181c-5p | 0.892 | 0.0134 | Resistant |
| miR-664b-3p | 0.733 | 0.0206 | - |
| miR-182-5p | 0.704 | 0.0134 | Susceptible |
| let-7d-5p | 0.464 | 0.0053 | Resistant |
| miR-181a-5p | 0.419 | 0.0422 | - |
| miR-15b-3p | −0.810 | 0.0008 | - |
| miR-494-3p | −0.961 | 0.0326 | Resistant |
| IFN-β | |||
| miR-450b-5p | −1.441 | 0.0020 | Resistant |
| IFN-γ | |||
| miR-664b-3p | 0.733 | 0.0206 | - |
| miR-26b-5p | −0.475 | 0.0313 | - |
| miR-374c-5p | −1.228 | 0.0096 | - |
| log2FC | padj | |
|---|---|---|
| miR-140-3p | −0.007953398 | 0.974817048 |
| miR-32-3p | 0.013920456 | 0.974817048 |
| miR-374a-5p | 0.020538776 | 0.945449302 |
| miR-196b-5p | 0.025080141 | 0.946019864 |
| miR-339-5p | 0.029116789 | 0.949059007 |
| miR-3158-3p | 0.029234925 | 0.97722196 |
| miR-320c | 0.033177006 | 0.970481801 |
| miR-423-3p | 0.039739622 | 0.860137595 |
| miR-331-5p | 0.04015155 | 0.957965014 |
| miR-1180-3p | 0.042520619 | 0.897231993 |
| miR-1229-3p | 0.049456491 | 0.979826711 |
| miR-6803-3p | 0.069110586 | 0.956513365 |
| miR-339-3p | 0.075904611 | 0.883114045 |
| miR-4326 | 0.149533463 | 0.815150225 |
| miR-16-2-3p | 0.157983774 | 0.530911494 |
| miR-6087 | 0.178211797 | 0.945394126 |
| miR-25-5p | 0.181172206 | 0.692047229 |
| miR-148b-3p | 0.20381122 | 0.369688028 |
| miR-126-3p | 0.20432634 | 0.517498738 |
| miR-760 | 0.204547203 | 0.876649033 |
| miR-200b-5p | 0.206829005 | 0.813533675 |
| miR-1307-5p | 0.221466282 | 0.90711043 |
| miR-454-3p | 0.226527458 | 0.372951464 |
| miR-107 | 0.232189082 | 0.645482953 |
| miR-301a-3p | 0.234614774 | 0.88095324 |
| miR-200b-3p | 0.241301831 | 0.267692191 |
| miR-324-3p | 0.254720466 | 0.852842736 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pierce, J.B.; Simion, V.; Icli, B.; Pérez-Cremades, D.; Cheng, H.S.; Feinberg, M.W. Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs. Genes 2020, 11, 1354. https://doi.org/10.3390/genes11111354
Pierce JB, Simion V, Icli B, Pérez-Cremades D, Cheng HS, Feinberg MW. Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs. Genes. 2020; 11(11):1354. https://doi.org/10.3390/genes11111354
Chicago/Turabian StylePierce, Jacob B., Viorel Simion, Basak Icli, Daniel Pérez-Cremades, Henry S. Cheng, and Mark W. Feinberg. 2020. "Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs" Genes 11, no. 11: 1354. https://doi.org/10.3390/genes11111354