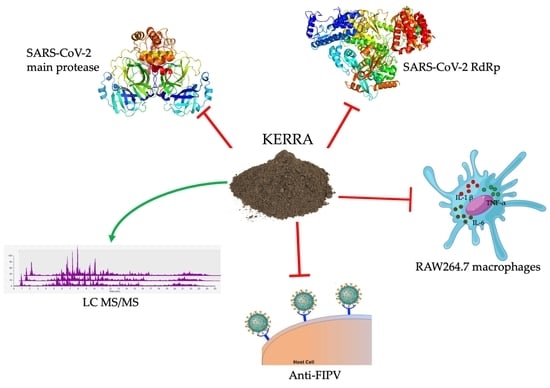
KERRA, Mixed Medicinal Plant Extracts, Inhibits SARS-CoV-2 Targets Enzymes and Feline Coronavirus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Inhibition of the Main Protease of SARS-CoV-2
2.2. SARS-CoV-2 RNA-Dependent RNA Polymerase Inhibition of KERRA
2.3. Anti-Inflammatory Effect of KERRA in Lipopolysaccharide-Stimulated RAW264.7 Macrophages
2.4. Anti-FIPV Activity Assay
2.5. Phytochemical Profile Analysis Using LC–MS/MS
3. Results
3.1. Inhibition of the Main Protease of SARS-CoV-2 by KERRA
3.2. KERRA Inhibition of RdRp (SARS-CoV-2)
3.3. Effect of KERRA on Anti-Inflammation Activity
3.4. Anti-FIPV Activity
3.5. Phytochemical Profiling and Qualitative Metabolite Analysis
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, H.; Stratton, C.W.; Tang, Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J. Med. Virol. 2020, 92, 401–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef]
- Singh, A.K.; Gupta, R.; Ghosh, A.; Misra, A. Diabetes in COVID-19: Prevalence, pathophysiology, prognosis and practical considerations. Diabetes Metab. Syndr. 2020, 14, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Patidar, R.; Younis, K.; Desai, P.; Hosein, Z.; Padda, I.; Mangat, J.; Altaf, M. Comorbidity and its Impact on Patients with COVID-19. SN Compr. Clin. Med. 2020, 2, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Priyanka, O.P.C.; Singh, I.; Patra, G. Aerosol transmission of SARS-CoV-2: The unresolved paradox. Travel Med. Infect. Dis. 2020, 37, 101869. [Google Scholar] [CrossRef]
- Priyanka, O.P.C.; Singh, I. Protective immunity against COVID-19: Unravelling the evidences for humoral vs. cellular components. Travel Med. Infect. Dis. 2021, 39, 101911. [Google Scholar] [CrossRef]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.C.; Yang, W.H.; Yang, C.S.; Hou, M.H.; Tsai, C.L.; Chou, Y.Z.; Hung, M.C.; Chen, Y. Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug. Am. J. Cancer Res. 2020, 10, 2535–2545. [Google Scholar]
- Razali, R.; Asis, H.; Budiman, C. Structure-Function Characteristics of SARS-CoV-2 Proteases and Their Potential Inhibitors from Microbial Sources. Microorganisms 2021, 9, 2481. [Google Scholar] [CrossRef]
- Jiang, Y.; Yin, W.; Xu, H.E. RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19. Biochem. Biophys. Res. Commun. 2021, 538, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Hillen, H.S. Structure and function of SARS-CoV-2 polymerase. Curr. Opin. Virol. 2021, 48, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Sa-Ngiamsuntorn, K.; Suksatu, A.; Pewkliang, Y.; Thongsri, P.; Kanjanasirirat, P.; Manopwisedjaroen, S.; Charoensutthivarakul, S.; Wongtrakoongate, P.; Pitiporn, S.; Chaopreecha, J.; et al. Anti-SARS-CoV-2 Activity of Andrographis paniculata Extract and Its Major Component Andrographolide in Human Lung Epithelial Cells and Cytotoxicity Evaluation in Major Organ Cell Representatives. J. Nat. Prod. 2021, 84, 1261–1270. [Google Scholar] [CrossRef]
- Daly, W.J. The black cholera comes to the central valley of America in the 19th century—1832, 1849, and later. Trans. Am. Clin. Climatol. Assoc. 2008, 119, 143–152. [Google Scholar] [PubMed]
- Sun, J.; Liu, J.N.; Fan, B.; Chen, X.N.; Pang, D.R.; Zheng, J.; Zhang, Q.; Zhao, Y.F.; Xiao, W.; Tu, P.F.; et al. Phenolic constituents, pharmacological activities, quality control, and metabolism of Dracaena species: A review. J. Ethnopharmacol. 2019, 244, 112138. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.W.; Wang, J.S.; Wang, Y.H.; Xiao, H.T.; Hu, X.J.; Mu, S.Z.; Ma, Y.L.; Lin, H.; He, H.P.; Li, L.; et al. Tarennane and tarennone, two novel chalcone constituents from Tarenna attenuata. Planta Med. 2007, 73, 496–498. [Google Scholar] [CrossRef]
- Rob, M.M.; Hossen, K.; Iwasaki, A.; Suenaga, K.; Kato-Noguchi, H. Phytotoxic Activity and Identification of Phytotoxic Substances from Schumannianthus dichotomus. Plants 2020, 9, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsoi, A.Y.; Ng, T.B.; Fong, W.P. Immunomodulatory activity of a chymotrypsin inhibitor from Momordica cochinchinensis seeds. J. Pept. Sci. 2006, 12, 605–611. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, W.; Cheng, L.; Li, M.; Huang, J.; Bao, S.; Xu, Q.; Ma, Z. Citrus fruits are rich in flavonoids for immunoregulation and potential targeting ACE2. Nat. Prod. Bioprospect. 2022, 12, 4. [Google Scholar] [CrossRef]
- Banskota, A.H.; Tezuka, Y.; Adnyana, I.K.; Xiong, Q.; Hase, K.; Tran, K.Q.; Tanaka, K.; Saiki, I.; Kadota, S. Hepatoprotective effect of Combretum quadrangulare and its constituents. Biol. Pharm. Bull. 2000, 23, 456–460. [Google Scholar] [CrossRef] [Green Version]
- Itthiarbha, A.; Phitak, T.; Sanyacharernkul, S.; Pothacharoen, P.; Pompimon, W.; Kongtawelert, P. Polyoxypregnane glycoside from Dregea volubilis extract inhibits IL-1beta-induced expression of matrix metalloproteinase via activation of NF-kappaB in human chondrocytes. In Vitro Cell. Dev. Biol. Anim. 2012, 48, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Sureram, S.; Senadeera, S.P.; Hongmanee, P.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. Antimycobacterial activity of bisbenzylisoquinoline alkaloids from Tiliacora triandra against multidrug-resistant isolates of Mycobacterium tuberculosis. Bioorg. Med. Chem. Lett. 2012, 22, 2902–2905. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Dwivedee, B.P.; Bisht, D.; Dash, A.K.; Kumar, D. The chemical constituents and diverse pharmacological importance of Tinospora cordifolia. Heliyon 2019, 5, e02437. [Google Scholar] [CrossRef] [Green Version]
- Ihssen, J.; Faccio, G.; Yao, C.; Sirec, T.; Spitz, U. Fluorogenic in vitro activity assay for the main protease M(pro) from SARS-CoV-2 and its adaptation to the identification of inhibitors. STAR Protoc. 2021, 2, 100793. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Echeverria, C.; Rich, D.H. New intramolecularly quenched fluorogenic peptide substrates for the study of the kinetic specificity of papain. FEBS Lett. 1992, 297, 100–102. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Chen, L.L.; Luo, H.B.; Sun, T.; Chen, J.; Ye, F.; Cai, J.H.; Shen, J.K.; Shen, X.; Jiang, H.L. Enzymatic activity characterization of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer technique. Acta Pharmacol. Sin. 2005, 26, 99–106. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Wu, J.; Wang, H.; Gao, Y.; Liu, Q.; Mu, A.; Ji, W.; Yan, L.; Zhu, Y.; Zhu, C.; et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell 2020, 182, 417–428.e13. [Google Scholar] [CrossRef]
- Yingchutrakul, Y.; Sittisaree, W.; Mahatnirunkul, T.; Chomtong, T.; Tulyananda, T.; Krobthong, S. Cosmeceutical Potentials of Grammatophyllum speciosum Extracts: Anti-Inflammations and Anti-Collagenase Activities with Phytochemical Profile Analysis Using an Untargeted Metabolomics Approach. Cosmetics 2021, 8, 116. [Google Scholar] [CrossRef]
- Huff, S.; Kummetha, I.R.; Tiwari, S.K.; Huante, M.B.; Clark, A.E.; Wang, S.; Bray, W.; Smith, D.; Carlin, A.F.; Endsley, M.; et al. Discovery and Mechanism of SARS-CoV-2 Main Protease Inhibitors. J. Med. Chem. 2022, 65, 2866–2879. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Huang, Y.Y.; Wu, Y.; Liu, R.; Zhou, L.; Lin, Y.; Wu, D.; Zhang, L.; Liu, H.; et al. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc. Natl. Acad. Sci. USA 2020, 117, 27381–27387. [Google Scholar] [CrossRef]
- Amporndanai, K.; Meng, X.; Shang, W.; Jin, Z.; Rogers, M.; Zhao, Y.; Rao, Z.; Liu, Z.J.; Yang, H.; Zhang, L.; et al. Inhibition mechanism of SARS-CoV-2 main protease by ebselen and its derivatives. Nat. Commun. 2021, 12, 3061. [Google Scholar] [CrossRef] [PubMed]
- Drozdzal, S.; Rosik, J.; Lechowicz, K.; Machaj, F.; Kotfis, K.; Ghavami, S.; Los, M.J. FDA approved drugs with pharmacotherapeutic potential for SARS-CoV-2 (COVID-19) therapy. Drug Resist. Updat. 2020, 53, 100719. [Google Scholar] [CrossRef] [PubMed]
- Magro, P.; Zanella, I.; Pescarolo, M.; Castelli, F.; Quiros-Roldan, E. Lopinavir/ritonavir: Repurposing an old drug for HIV infection in COVID-19 treatment. Biomed. J. 2021, 44, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Group, R.C. Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2020, 396, 1345–1352. [Google Scholar] [CrossRef]
- Ma, C.; Sacco, M.D.; Hurst, B.; Townsend, J.A.; Hu, Y.; Szeto, T.; Zhang, X.; Tarbet, B.; Marty, M.T.; Chen, Y.; et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ma, C.; Tan, H.; Choza, J.; Wang, Y.; Wang, J. Validation and invalidation of SARS-CoV-2 main protease inhibitors using the Flip-GFP and Protease-Glo luciferase assays. Acta Pharm. Sin. B 2022, 12, 1636–1651. [Google Scholar] [CrossRef]
- Wang, X.; Sacramento, C.Q.; Jockusch, S.; Chaves, O.A.; Tao, C.; Fintelman-Rodrigues, N.; Chien, M.; Temerozo, J.R.; Li, X.; Kumar, S.; et al. Combination of antiviral drugs inhibits SARS-CoV-2 polymerase and exonuclease and demonstrates COVID-19 therapeutic potential in viral cell culture. Commun. Biol. 2022, 5, 154. [Google Scholar] [CrossRef]
- Driouich, J.S.; Cochin, M.; Lingas, G.; Moureau, G.; Touret, F.; Petit, P.R.; Piorkowski, G.; Barthelemy, K.; Laprie, C.; Coutard, B.; et al. Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model. Nat. Commun. 2021, 12, 1735. [Google Scholar] [CrossRef]
- Jin, Y.H.; Min, J.S.; Jeon, S.; Lee, J.; Kim, S.; Park, T.; Park, D.; Jang, M.S.; Park, C.M.; Song, J.H.; et al. Lycorine, a non-nucleoside RNA dependent RNA polymerase inhibitor, as potential treatment for emerging coronavirus infections. Phytomedicine 2021, 86, 153440. [Google Scholar] [CrossRef]
- Min, J.S.; Kwon, S.; Jin, Y.H. SARS-CoV-2 RdRp Inhibitors Selected from a Cell-Based SARS-CoV-2 RdRp Activity Assay System. Biomedicines 2021, 9, 996. [Google Scholar] [CrossRef]
- Fan, S.; Xiao, D.; Wang, Y.; Liu, L.; Zhou, X.; Zhong, W. Research progress on repositioning drugs and specific therapeutic drugs for SARS-CoV-2. Future Med. Chem. 2020, 12, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Furuta, Y.; Komeno, T.; Nakamura, T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 449–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woranam, K.; Senawong, G.; Utaiwat, S.; Yunchalard, S.; Sattayasai, J.; Senawong, T. Anti-inflammatory activity of the dietary supplement Houttuynia cordata fermentation product in RAW264.7 cells and Wistar rats. PLoS ONE 2020, 15, e0230645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maia, M.; Resende, D.; Duraes, F.; Pinto, M.M.M.; Sousa, E. Xanthenes in Medicinal Chemistry—Synthetic strategies and biological activities. Eur. J. Med. Chem. 2021, 210, 113085. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.N.; Trinite, B.; Levy, D.N. Potent Inhibition of HIV-1 Replication in Resting CD4 T Cells by Resveratrol and Pterostilbene. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [Green Version]
- Markowicz, J.; Uram, L.; Sobich, J.; Mangiardi, L.; Maj, P.; Rode, W. Antitumor and anti-nematode activities of alpha-mangostin. Eur. J. Pharmacol. 2019, 863, 172678. [Google Scholar] [CrossRef]
- Yeo, S.C.M.; Fenwick, P.S.; Barnes, P.J.; Lin, H.S.; Donnelly, L.E. Isorhapontigenin, a bioavailable dietary polyphenol, suppresses airway epithelial cell inflammation through a corticosteroid-independent mechanism. Br. J. Pharmacol. 2017, 174, 2043–2059. [Google Scholar] [CrossRef]
- Fang, J.; Wu, Q.; Ye, F.; Cai, C.; Xu, L.; Gu, Y.; Wang, Q.; Liu, A.L.; Tan, W.; Du, G.H. Network-Based Identification and Experimental Validation of Drug Candidates Toward SARS-CoV-2 via Targeting Virus-Host Interactome. Front. Genet. 2021, 12, 728960. [Google Scholar] [CrossRef]
- Yancey, P.H. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; He, F.; Wu, C.; Li, P.; Li, N.; Deng, J.; Zhu, G.; Ren, W.; Peng, Y. Betaine in Inflammation: Mechanistic Aspects and Applications. Front. Immunol. 2018, 9, 1070. [Google Scholar] [CrossRef] [Green Version]






| List | Name | Formula | Molecular Weight (Da) | Area |
|---|---|---|---|---|
| 1 | 2-Methoxy-9H-xanthen-9-one | C14 H10 O3 | 226.0594 | 7.43 × 109 |
| 2 | Isorhapontigenin | C15 H14 O4 | 276.0960 | 7.36 × 109 |
| 3 | Betaine | C5 H11 N O2 | 117.0776 | 6.84 × 109 |
| 4 | - | C20 H28 O4 | 314.1837 | 6.81 × 109 |
| 5 | trans-Anethole | C10 H12 O | 148.0869 | 4.82 × 109 |
| 6 | Eicosatetraynoic acid | C20 H24 O2 | 296.1734 | 4.05 × 109 |
| 7 | NP-020078 | C17 H28 O3 | 302.1847 | 3.28 × 109 |
| 8 | NP-003294 | C18 H16 O7 | 344.0850 | 3.09 × 109 |
| 9 | - | C20 H30 O5 | 332.1946 | 2.88 × 109 |
| 10 | N1-(3-chlorophenyl)-2-[2-(trifluoromethyl)-4-quinolyl]hydrazine-1-carboxamide | C17 H12 Cl F3 N4 O | 380.0672 | 2.37 × 109 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seetaha, S.; Khamplong, P.; Wanaragthai, P.; Aiebchun, T.; Ratanabunyong, S.; Krobthong, S.; Yingchutrakul, Y.; Rattanasrisomporn, J.; Choowongkomon, K. KERRA, Mixed Medicinal Plant Extracts, Inhibits SARS-CoV-2 Targets Enzymes and Feline Coronavirus. COVID 2022, 2, 621-632. https://doi.org/10.3390/covid2050046
Seetaha S, Khamplong P, Wanaragthai P, Aiebchun T, Ratanabunyong S, Krobthong S, Yingchutrakul Y, Rattanasrisomporn J, Choowongkomon K. KERRA, Mixed Medicinal Plant Extracts, Inhibits SARS-CoV-2 Targets Enzymes and Feline Coronavirus. COVID. 2022; 2(5):621-632. https://doi.org/10.3390/covid2050046
Chicago/Turabian StyleSeetaha, Supaphorn, Phatcharin Khamplong, Panatda Wanaragthai, Thitinan Aiebchun, Siriluk Ratanabunyong, Sucheewin Krobthong, Yodying Yingchutrakul, Jatuporn Rattanasrisomporn, and Kiattawee Choowongkomon. 2022. "KERRA, Mixed Medicinal Plant Extracts, Inhibits SARS-CoV-2 Targets Enzymes and Feline Coronavirus" COVID 2, no. 5: 621-632. https://doi.org/10.3390/covid2050046