Abstract
By targeting the essential viral RNA-dependent RNA polymerase (RdRP), nucleoside analogs (NAs) have exhibited great potential in antiviral therapy for RNA virus-related diseases. However, most ribose-modified NAs do not present broad-spectrum features, likely due to differences in ribose-RdRP interactions across virus families. Here, we show that HNC-1664, an adenosine analog with modifications both in ribose and base, has broad-spectrum antiviral activity against positive-strand coronaviruses and negative-strand arenaviruses. Importantly, treatment with HNC-1664 demonstrate anti-SARS-CoV-2 efficacy in infected K18-human ACE2 mice, with reduced viral titer and mortality, as well as improved lung injury. Enzymology data demonstrate that HNC-1664 inhibits RNA synthesis mainly at the pre-catalysis stage. The cryo-EM structures of HNC-1664-bound RdRP-RNA complexes from both SARS-CoV-2 and LASV reveal an unusual base pairing mode of HNC-1664 in part due to its base modification, thus revealing its great potency in binding but not catalysis. Under certain circumstances, 1664-TP can be slowly incorporated by RdRP through regular Watson-Crick base pairing, as evidenced by enzymology data and an HNC-1664-incorporated crystal structure of the RdRP-RNA complex. Overall, HNC-1664 achieves broad-spectrum characteristics by favoring an alternative base pairing strategy to non-catalytically block RNA synthesis, providing a novel concept for the rational development of NA drugs.
Similar content being viewed by others
Introduction
Infections caused by RNA viruses present a significant challenge to public health. In addition, owing to the high frequency of mutations and recombinations in RNA genomes, several RNA viruses cause continuous threats1. Over the past two decades, coronaviruses (with a positive-sense single-stranded RNA genome) have repeatedly emerged and caused epidemics or pandemics in humans, including severe acute respiratory syndrome coronavirus (SARS-CoV) during 2002–20032, Middle East respiratory syndrome coronavirus (MERS-CoV) in 20123, and the ongoing prevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) since the end of 20194. In particular, the emergence of SARS-CoV-2 variants of concern (VOCs), such as XBB.1.18, HK.3.1, and BF.7.14, has increased the reinfection rate and persistent threat of SARS-CoV-2 infection to global public health5,6. Furthermore, among other related zoonotic groups of coronaviruses, the Alphacoronavirus HCoV-229E and HCoV-NL63, along with the Betacoronavirus HCoV-OC43 and HCoV-HKU1, can infect humans and usually result in mild upper respiratory tract infection, with symptoms similar to those of the common cold, such as fatigue and a runny nose7. Recent studies have discovered several compounds that exhibit broad-spectrum antiviral activity, not only against positive-strand RNA viruses but also against various negative-strand RNA viruses, including arenaviruses8. Several members of the arenavirus family cause severe hemorrhagic fever syndrome in humans9. LASV, in particular, leads to Lassa fever (LF), which is endemic in West Africa and has recently caused outbreaks in several West African countries (https://ncdc.gov.ng/diseases/sitreps)10. Compared with LASV, lymphocytic choriomeningitis virus (LCMV)-induced disease is rarely as severe as LF. Nonetheless, LCMV is widely distributed across the globe and causes neurological disorders in some cases. Accordingly, LCMV is commonly used as an authentic model of biosafety level 4 (BSL-4) pathogenic LASV under BSL-2 conditions11,12. Currently, there are no approved antiviral drugs or vaccines for LASV or LCMV. Diseases caused by SARS-CoV-2, SARS-CoV, MERS-CoV, and LASV have been designated by the World Health Organization (WHO) as priority diseases for research and development in emergency contexts. Identification of the common mechanism or targets involved in the life cycle of various RNA viruses is believed to be an ideal strategy for discovering pan-RNA virus inhibitors. Therefore, this study aimed to determine the common targets involved and identify potential broad-spectrum inhibitors to address the challenges of the current epidemic and possible future outbreaks caused by different families of RNA viruses.
The virally encoded RNA-dependent RNA polymerase (RdRP) is essential for RNA viruses due to its responsibility for whole-genome replication and mRNA transcription13,14. Nucleoside/nucleotide analogs (NAs), which target viral RdRP, are ideal drug candidates with both high inhibition and broad-spectrum potentials15. However, very few NA drugs have been found to be clinically successful at treating diseases caused by RNA viruses. Among them, the hepatitis C drug sofosbuvir, a uridine analog with key modifications at the ribose 2′-position, is a prominent example that efficiently blocks hepatitis C virus (HCV) RNA synthesis, likely through immediate chain termination after incorporation into the product RNA chain16,17. Remdesivir, an adenine analog with key modifications at the ribose 1′-position, was originally developed to combat Ebola virus and eventually approved as an anti-SARS-CoV-2 drug for emergency use, causing a steric clash by the 1′-cyano group with the side chain of S861 upstream of the RdRP catalytic site, resulting in delayed intervention upon the third nucleotide addition cycle after its incorporation18,19,20. Thus, ribose-modified NAs can possibly block RNA synthesis either at the incorporation step or after incorporation. However, their broad-spectrum potential has not been adequately demonstrated. Upon binding and catalysis, NTP ribose interacts mainly with motifs A-C21,22, the most conserved parts of the viral RdRP. However, at the entire RNA virus group level, motifs A-C contain only three invariant residues, one in each motif23. Hence, a ribose-modified compound likely induces interactions with subtle differences across different RdRPs. In contrast, base-modified NAs, such as favipiravir and molnupiravir, can establish Watson-Crick-type base pairing interactions with templating nucleotides in a relatively conservative manner and therefore show much broader spectrum features than ribose-modified NAs do24,25. However, the high mutagenic potential of favipiravir and molnupiravir is of concern26,27. To prepare for emerging RNA viruses, the discovery of more NAs with both broad-spectrum features and unique modifications is critical.
In this work, the adenosine analog HNC-1664 (HNC; Henovcom Bioscience Co., Ltd.) modified with both ribose and base moieties is shown to be highly effective against both positive-strand coronaviruses and negative-strand arenaviruses. With the nomenclature (2R,3R,4S,5R)−4-fluoro-2-(hydroxymethyl)−5-(5-iodo-4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl) tetrahydrofuran-3-ol (Fig. 1), HNC-1664 is among a series of 7-deazapurine nucleoside derivatives developed as potential anti-dengue virus agents (named compound 13b in the original publication)28. RdRP enzymatic data from SARS-CoV-2 and LASV indicate that HNC-1664 inhibits RNA synthesis mainly in a noncatalytic manner. Cryo-electron microscopy (cryo-EM) data from SARS-CoV-2 (3.3 Å resolution) and LASV (3.0 Å resolution) RdRP-RNA-HNC-1664 complexes demonstrate that this rarely occurring intervention mode is achieved through a noncanonical base pairing interaction with the purine base flipped by nearly 180°. Consequently, RdRP cannot proceed to close the active site for catalysis. Under certain circumstances, this compound can be slowly incorporated by SARS-CoV-2 and mechanistically related enterovirus 71 (EV71) RdRPs via primer-dependent initiation mechanisms. The crystal structure of the EV71 RdRP-RNA-HNC-1664 complex solved at 3.0 Å resolution further demonstrate that such incorporation was likely achieved through regular Watson-Crick base pairing. In summary, we identify an NA molecule that has broad-spectrum effects on different families of RNA viruses and elucidate the comprehensive mechanisms underlying its effects on viral RdRPs.
a Representative IFA images showing the SARS-CoV-2 NP protein (green) and nuclei (blue) in Vero E6 cells (n = 3). b Dose-response curves of HNC-1664 for the inhibition of infection (solid black line) and cell viability (dashed blue line) in Vero E6 cells (n = 3). c Representative IFA images showing the SARS-CoV-2 WT strain NP protein (green) and nuclei (blue) in Caco-2 cells (n = 3). d Dose-response curves of HNC-1664 (solid red line) and remdesivir (dashed black line) for the inhibition of SARS-CoV-2 WT strain infection in Caco-2 cells (n = 3). Dose-response curve of cell viability (dashed blue line) was evaluated via the CCK-8 assay (n = 6). e–g Dose-response curves of HNC-1664 (solid red line) and remdesivir (dashed black line) for the inhibition of XBB.1.18.1 (e), HK.3.1 (f), and BF.7.14 (g) variant infection (n = 3). h, i Antiviral activity of HNC-1664 against HCoV-229E (h) and HCoV-OC43 (i). Infected cells were collected and lysed at 24 h.p.i. for viral RNA detection via RT–qPCR (n = 3). The quantification of viral RNA was normalized to the vehicle control group. j Dose-response curves of HNC-1664 for cell viability in Huh-7 cells (dashed blue line) and RD cells (solid blue line). Cell viability was evaluated via the CCK-8 assay (n = 4). k IC50 values for HNC-1664 and remdesivir. The percentage of inhibition and cell viability was calculated relative to the vehicle control group. IC50 values were calculated using GraphPad Prism version 8.0 and are presented as the mean ± SD of three independent experiments. All data are presented as the mean ± SD. Statistical significance was assessed by one-way ANOVA with Dunnett’s post-hoc test compared with the vehicle control group. **P < 0.01, ***P < 0.001, ****P < 0.0001; ns no significance.
Results
Antiviral effects of HNC-1664 against SARS-CoV-2 and other human-pathogenic coronaviruses
To evaluate the antiviral effects of HNC-1664, we used Vero E6 cells to determine the half-maximal inhibitory concentration (IC50). First, Vero E6 cells were challenged with the SARS-CoV-2 wild-type (WT) strain. The data revealed that the antiviral effects of HNC-1664 against the SARS-CoV-2 WT strain were dose dependent, with an IC50 of 0.029 μM, and the expression of the viral nucleocapsid protein (NP) was rarely detectable in 0.1 μM HNC-1664-treated cells (Fig. 1a, b). Importantly, the cytotoxicity of HNC-1664 was tested via a CCK-8 assay, which revealed relatively weak cytotoxicity even at a concentration of 10 μM (Fig. 1b). To verify these results, we investigated the antiviral activity of HNC-1664 in a human colorectal cancer cell line (Caco-2), which is more physiologically related. Similarly, HNC-1664 robustly inhibited SARS-CoV-2 replication in Caco-2 cells, with an IC50 value of 0.076 μM and a 90.28% reduction in the viral protein concentration at 10 μM, whereas the cytotoxicity to related cells was only 10 ~ 20% (Fig. 1c, d). To further explore the antiviral effects of HNC-1664 against ongoing SARS-CoV-2 variants of concern (VOCs), we subsequently performed IFA (immunofluorescence assay) for the XBB.1.18.1, HK.3.1 and BF.7.14 variants in Vero E6 cell lines. HNC-1664 robustly inhibited viral production, with IC50 values of 0.37 μM, 1.14 μM, and 0.12 μM, respectively (Fig. 1e–g). Furthermore, we evaluated the antiviral efficacy of HNC-1664 against prevalent emerging VOCs, including alpha, beta, delta and Omicron BA.1. As expected, our data demonstrated that HNC-1664 exhibited high potency against these variants, with IC50 values of 0.019 μM, 0.064 μM, 0.084 μM and 0.096 μM, respectively (Supplementary Fig. 1a–e). To compare with remdesivir, we similarly tested the antiviral activitiy of remdesivir. The results indicated that the antiviral efficacy of HNC-1664 was comparable to that of remdesivir (Fig. 1d–g and Supplementary Fig. 1a–e).
The protein sequence of RdRP is highly similar (≥75%) among coronaviruses29. To further explore the antiviral effects of HNC-1664 on other seasonal coronaviruses, we investigated the effects of HNC-1664 against HCoV-229E and HCoV-OC43 on Huh-7 and RD cells. As shown in Fig. 1h, i, HNC-1664 reduced HCoV-229E and HCoV-OC43 replication in a dose-dependent manner. When HNC-1664 was used at a concentration of 1 μM, viral gene production decreased by approximately 92% in Huh-7 cells infected with HCoV-229E and by approximately 67% in RD cells infected with HCoV-OC43 (Fig. 1h, i). The cytotoxic effects of HNC-1664 on Huh-7 and RD cells were further determined. HNC-1664 exhibited modest cytotoxicity at a concentration corresponding to high inhibitory activity and showed ~40% cytotoxicity in both Huh-7 and RD cells (Fig. 1j). Notably, the antiviral effects on HCoV-229E and HCoV-OC43 were comparable to those of HNC-1664 on SARS-CoV-2 (Fig. 1k). Taken together, our results demonstrated that HNC-1664 has pan-coronavirus antiviral activity.
Broad-spectrum antiviral effects of HNC-1664 against arenaviruses
To explore the broad-spectrum antiviral effects of HNC-1664 against arenaviruses, we first investigated its antiviral activity against LASV by using the LASV minigenome (mini) system under biosafety level 2 (BSL-2) conditions. As shown in Fig. 2a, HNC-1664 inhibited the gene expression of LASV mini in a dose-dependent manner. After treatment with HNC-1664 for 24 h, the luciferase activity was reduced by approximately 87%, 66%, and 36% at concentrations of 10, 1, and 0.1 μM, respectively. Moreover, ribavirin was used as a positive control at a concentration of 250 μM. To consolidate these results, LCMV was employed as an authentic representative virus of arenaviruses and is closely related to the BSL-4 pathogenic LASV in genetics. Initially, we performed the same experiment on the LCMV minigenome system, and at concentrations of 10, 1, and 0.1 μM, HNC-1664 strongly inhibited the gene expression of LCMV mini, reducing luciferase activity by approximately 84%, 72%, and 51%, respectively (Fig. 2b). Overall, HNC-1664 showed comparable activity against LASV and LCMV mini. To confirm these results, the cytotoxic effects of HNC-1664 on Vero E6 and HEK293T cells transfected with the minigenome system were determined in parallel. Compared with that of non-transfected cells, the cytotoxicity of HNC-1664 did not differ at the tested concentrations (Supplementary Fig. 1f–h). The antiviral effects of HNC-1664 against the LCMV Armstrong strain in infected Huh-7 cells were subsequently determined, and the IC50 value for Huh-7 was 0.90 μM. Notably, the LCMV NP protein was almost undetected at a concentration of 10 μM (Fig. 2c, d). In summary, HNC-1664 has potent broad-spectrum antiviral effects on different families of RNA viruses, including coronaviruses and arenaviruses.
a Luciferase activity of the LASV minigenome after treatment with HNC-1664 (n = 5). Vero E6 cells were cotransfected with LASV mini, LASV NP and L helper plasmids. Luciferase activity was normalized to the vehicle control group. b Luciferase activity of the LCMV minigenome treated with HNC-1664 (n = 5). HEK293T cells were cotransfected with LCMV mini, LCMV NP and L helper plasmids. The procedures for collecting and processing the data are similar to those in panel (a). c Antiviral activity of HNC-1664 against the LCMV Armstrong strain. Representative IFA images showing the LCMV NP protein (green) and nuclei (blue) in Huh-7 cells (n = 3). d Dose-response curve of HNC-1664 for the inhibition of the LCMV Armstrong strain. The data were analyzed by IFA (n = 3). The inhibitory effect of HNC-1664 was calculated relative to the vehicle control group. The IC50 value is presented as the mean ± SD of three independent experiments. All data are presented as the mean ± SD. Statistical significance was assessed by one-way ANOVA with Dunnett’s post-hoc test compared with the vehicle control group. ***P < 0.001, ****P < 0.0001; ns no significance.
In vivo antiviral efficacy of HNC-1664 in K18-hACE2 transgenic mice
To verify the antiviral efficacy of HNC-1664 in vivo, we utilized the K18-human angiotensin-converting enzyme 2 (K18-hACE2) transgenic mouse model infected with the SARS-CoV-2 delta strain, which supports robust SARS-CoV-2 replication with lethal infection. A 50 mg kg−1 dose of HNC-1664 or 500 mg kg−1 dose of molnupiravir was administered by oral gavage to the mice starting at 2 h post-infection (h.p.i.). Similarly, the animals in the control group were treated with vehicle (Fig. 3a). HNC-1664 treatment protected the mice from SARS-CoV-2 delta-related lethal infection, as did molnupiravir treatment (HNC-1664 treatment: 7 of 7 or 100% vs molnupiravir treatment: 6 of 6 or 100% vs control group: 4 of 6 or 66.7%) (Fig. 3b). In addition, the production of infectious virus particles in mouse tissues was inhibited by the administration of HNC-1664, resulting in a decrease in viral titer by 33.8-fold in the nasal turbinate, 47.6-fold in the trachea, and 786.7-fold in the lungs (Fig. 3c–e). Consistent with the findings of the viral titer, the number of viral gene copies in the nasal turbinate, trachea, and lungs of HNC-1664-treated mice was markedly lower than that in the vehicle control group, with a reduction in viral gene copies ranging from 134.8- to 370.7-fold (Fig. 3f–h). In comparison, we measured the same viral replication indicators in molnupiravir-treated mice. Treatment with molnupiravir decreased the number of infectious virus particles and viral gene copies in the tissues (Fig. 3c–h). Importantly, the 50 mg kg−1 dose of HNC-1664 resulted in the clearance of infectious virus and viral gene copies in the lungs of 6 of the 7 mice, which was much lower than that resulting from the dose of 500 mg kg−1 dose molnupiravir (Fig. 3e, h).
a Schematic flow chart of the experimental design. K18-hACE2 transgenic mice were challenged with 1 × 105 PFU of the SARS-CoV-2 delta strain. Starting at 2 h post infection (h.p.i.), each mouse was administered HNC-1664 (n = 7), molnupiravir (n = 6) or vehicle (n = 6) by oral gavage twice daily. The image elements were created with BioRender. b Mouse survival was monitored daily until 4 days post infection (d.p.i.). The data are shown as Kaplan-Meier survival curves. c–e The number of infectious virus particles in the nasal turbinate (c), trachea (d) and lungs (e) of infected mice from the HNC-1664 (n = 7), molnupiravir (n = 6) and vehicle (n = 5) groups was determined via plaque assay. f–h The number of viral genes in the nasal turbinate (f), trachea (g) and lungs (h) of infected mice from the HNC-1664 (n = 7), molnupiravir (n = 6) and vehicle (n = 5) groups was determined via qRT-PCR. i, j HNC-1664 decreased damage in infected mouse lung tissue. Representative images of immunohistochemical staining (i) showing the presence of the SARS-CoV-2 NP protein (brown) in the lungs of infected mice at 4 d.p.i. Scale bar, 200 µm. Representative images of hematoxylin and eosin (H&E) staining (j) showing virus-induced damage in the lungs of infected mice at 4 d.p.i. Scale bar, 200 µm. The alveoli, bronchioles, and blood vessels of the lung were enlarged, and representative images are shown with enlarged images ①, ②, and ③, respectively. Scale bar, 50 µm. The data are presented as the mean ± SD (c–h). Statistical significance was assessed by one-way ANOVA with Dunnett’s post-hoc test compared with the vehicle control group. **P < 0.01, ***P < 0.001, ****P < 0.0001. LOD limit of detection.
Next, immunohistochemical staining revealed abundant expression of the SARS-CoV-2 nucleocapsid protein (NP) in the lungs of vehicle-treated mice (Fig. 3i, top). Consistent with the virological results, the expression of viral NP was hardly detected in the HNC-1664-treated group (Fig. 3i, middle) or the molnupiravir-treated group (Fig. 3i, bottom). To investigate whether HNC-1664 treatment contributes to alleviating histological injury caused by viral disease in K18-hACE2 mice, hematoxylin-eosin (H&E) staining was performed to assess virus-induced tissue damage in the lungs (Fig. 3j). In the lungs of the vehicle-treated mice, alveolar interstitial collapse was frequently observed in the alveoli, and alveolar wall thickening and the disappearance of alveolar septa were observed (Fig. 3j, top, enlarged image ①). Around the bronchioles, a necrotic epithelium, alveolar congestion and severe inflammatory infiltration were detected (Fig. 3j, top, enlarged image ②). Moreover, in blood vessels, congestion and mononuclear cell infiltration were frequently observed (Fig. 3j, top, enlarged image ③). In contrast, overall histological injury in mice receiving HNC-1664 or molnupiravir treatment was markedly suppressed (Fig. 3j, middle and bottom, enlarged images ①–③). Taken together, our data demonstrated that treatment with HNC-1664 potently protected K18-hACE2 mice against lethal SARS-CoV-2 delta variant infection via in vivo antiviral efficacy and suppressed viral production and virus-induced pathological injury in tissues.
HNC-1664 can inhibit viral RNA synthesis at the pre-catalysis stage through an unusual base pairing mechanism
To understand the intervention mechanisms of HNC-1664 at the RdRP level, we synthesized the NTP form of HNC-1664 (abbreviated as 1664-TP) and characterized its effect on the SARS-CoV-2 RdRP system. In accordance with the overall structures of ATP and 1664-TP, 1664-TP was modified at its ribose 2′-position with fluoro and its base with 6-methyl and 7-iodo moieties (Fig. 4a, b). A minimal 37-mer RNA hairpin substrate construct containing a FAM fluorophore labeled on its 5′ end was used to allow the incorporation of AMP at five consecutive incorporation sites (Fig. 4c, top). When SARS-CoV-2 nonstructural protein 12 (nsp12), nsp7, and nsp8 were incubated at a molar ratio of 1:1:2, the RdRP holoenzyme formed (Supplementary Fig. 2a–c), which activated the RdRP module within nsp12 for RNA synthesis30,31. The RdRP holoenzyme was able to incorporate AMP and extend the primer by five nucleotides when ATP or a mixture of all four NTPs was offered at a concentration of 0.25 mM or 0.25 mM each (Fig. 4c, bottom, lanes 2–4 and Supplementary Fig. 2d, e). In the presence of 0.25 mM ATP, 1664-TP at 0.05 mM and 0.1 mM still allowed AMP incorporation but reduced the level by 26–71% (Fig. 4c, bottom, lanes 5–10). When the concentration of 1664-TP was increased to 1 mM, no AMP incorporation was observed, even after 60 min (Fig. 4c, bottom, lanes 11–13). Typically, catalytically competent NA drugs can cause intervention at the nucleotide addition stage or after incorporation. However, 1664-TP likely intervenes in the pre-catalysis stages. Unlike primer-dependent SARS-CoV-2 RdRP, Bunyavirales RdRPs, such as the LASV L protein, can initiate RNA synthesis de novo or through short primers of 2–3 nucleotides in length. Using a 30-mer template (T30) and a trinucleotide primer (P3) as a combination, the LASV L protein can incorporate an AMP at the first incorporation site (Fig. 4d, top). Unlike SARS-CoV-2 nsp12, which requires both nsp7 and nsp8 for RdRP activity, the LASV L protein alone is an active RdRP. In the preparatory analysis, we compared a pair of RNA constructs with or without a downstream hairpin and found that the construct with the downstream hairpin resulted in a higher level of RNA synthesis than the other construct in trinucleotide-driven 9-mer product synthesis in the presence of ATP and UTP (Supplementary Fig. 3, comparing lanes 4–5 and 2–3), whereas the presence of the hairpin did not hinder extension of the 9-mer to the 10-mer when CTP was further supplied (Supplementary Fig. 3, comparing lanes 8–9 and 6–7). Hence, the downstream hairpin was retained in our construct design for LASV RdRP. When ATP was provided at a concentration of 0.25 mM, the L protein could incorporate AMP in a time-dependent manner (Fig. 4d, bottom, lanes 2–4). When 1664-TP was added at concentrations of 0.05 mM and 0.1 mM in the presence of 0.25 mM ATP, incorporation was still evident, but at the longest time point tested (60 min), the levels were reduced by 36% and 56%, respectively (Fig. 4d, bottom, compare lane 4 with lanes 7 and 10). When 1664-TP was added at a 1 mM concentration, the incorporation of AMP by the LASV L protein was completely abolished (Fig. 4d, bottom, lanes 11–13). Moreover, incorporation was not observed when 1664-TP was provided as the sole substrate at 0.25 mM (Fig. 4d, bottom, lanes 14–16). When the next incoming substrate(s) UTP or UTP/CTP were provided together with 1664-TP at 0.25 mM each, incorporation was still not observed (Supplementary Fig. 4). Taken together, 1664-TP exhibited an inhibitory effect but did not lead to incorporation under these conditions.
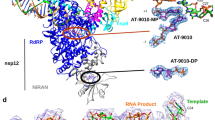
a, b Structural formulas of ATP (a) and 1664-TP (b). c 1664-TP inhibits SARS-CoV-2 RNA synthesis in a non-catalytic manner. Top: Schematic diagram of the minimal RNA substrate used for the SARS-CoV-2 incorporation assay. Bottom: Representative images showing the inhibitory effect of 1664-TP on SARS-CoV-2 RdRP replication. The amount of extended RNA at the longest time point tested (60 min) (lane 4) was used as the standard (set as 100) to quantify the extended RNA products. The reduction levels were calculated by comparing the lanes at the same time point tested (compare lane 2 with lanes 5 and 8 for 10 min, compare lane 3 with lanes 6 and 9 for 30 min, and compare lane 4 with lanes 7 and 10 for 60 min). Experiment was repeated three times independently with similar results. d 1664-TP inhibits LASV RNA synthesis in a non-catalytic manner. Top: The T30/P3 construct used for the assay. Bottom: Representative images showing the inhibition of LASV RdRP replication activity by 1664-TP. The amount of 4-mer in the marker lane (M, lane 1) was used as the standard (set as 100) to quantify the 4-mer products. Experiment was repeated twice independently with similar results. e Atomic model of 1664-TP binding with SARS-CoV-2 RdRP, viewed toward the edge of the 1664-TP purine base (left) and the 1664-TP purine plane (middle). Right: close contacts (4.1 Å or shorter in distance between nonhydrogen atoms) involving the 1664-TP base and ribose atoms are indicated by black dashed lines. Motifs A/B/F, each containing one invariant RdRP residue, are indicated by capital letters with a gray background. f Cryo-EM structure of a 1664-TP-bound LASV-RdRP-RNA complex. Top: Viewing toward the edge of the 1664-TP purine base; Bottom: Viewing toward the 1664-TP purine plane. Left: The overlay of the LASV-RNA-1664-TP structure with the EM density map (contoured at 6σ). Middle: The LASV-RNA-UMPNPP structure was used as a reference. Right: A superimposition of the 1664-TP- and UMPNPP-containing structures. The distances between the 1′-carbon atoms and the 4′-carbon atoms were used to estimate the positional difference between the NTP ribose moieties in these two structures. g Stereo pair images of the interaction details of 1664-TP with the LASV RdRP EC. The α-carbon atoms of the four invariant RdRP residues are shown as brown spheres. Close contacts (4.1 Å or shorter in distance between nonhydrogen atoms) involving the 1664-TP base and ribose atoms are indicated by purple dashed lines. Motifs A/B/C/F, each containing one invariant RdRP residue, are indicated by capital letters with a gray background.
To further reveal the structural basis of this unique intervention mode of 1664-TP, we solved 1664-TP-containing cryo-electron microscopy (cryo-EM) structures of both SARS-CoV-2 and LASV RdRP-RNA complexes via single-particle reconstruction methods (Supplementary Tables 1–2; Supplementary Figs. 5, 6, 7, and 8a, b). In both structures, 1664-TP occupies the active site through an alternative base pairing mode that was previously unobserved in the nucleic acid polymerase active site (Fig. 4e, f). The purine nucleoside still adopts the energetically favorable anti conformation, but the purine plane is almost flipped by 180° along the regular base pairing direction, resulting in a non-Watson-Crick base pairing feature by one hydrogen bond between the purine N1 and pyrimidine N3 atoms (Fig. 4e, f). Consequently, the placement of the 1664-TP ribose in the 3.0-Å LASV structure is 6–8 Å apart from the reference position in a UTP analog-bound LASV structure with a closed active site (Fig. 4f, bottom left). In the 3.3-Å SARS-CoV-2 structure, the conformation of the 1664-TP nucleoside is largely consistent with that in the LASV structure, except for the local orientation difference of the ribose due to the rotational movement along the purine N9-ribose 1′-C bond. Since the precise placement of NTP ribose 2′- and 3′-hydroxyls is critical for triggering nucleotide addition in viral RdRPs17,21,22, the alternative base pairing mode observed in both the SARS-CoV-2 and LASV RdRP structures explains the non-catalytic intervention feature of 1664-TP. Dissociation-and-rebinding is likely necessary for 1664-TP to switch between this unusual base pairing and the regular Watson-Crick base pairing with the templating nucleotide. In the SARS-CoV-2 catalytic center, the amidogens K545 and R555 can form close non-bonded contacts with the N5 atom of the base moiety of 1664-TP. This bonding arrangement is bolstered by positively charged residues from motif F (K551, K553, R555) and motif A (K621) surrounding the triphosphate moiety, creating a restrictive environment for 1664-TP and limiting the entry of new nucleotide substrates into the catalytic center (Fig. 4e, left). The details of the interaction between 1664-TP and LASV RdRP further clarify the binding potency of this compound (Fig. 4g and Supplementary Fig. 9). The purine base is stabilized by close contacts (approximately 4 Å or shorter in distance between non-hydrogen atoms) from motif B involving characteristic modifications of the 6-methyl and 7-iodo groups. The relocated ribose 3′-hydroxyl forms a hydrogen bond with the highly conserved motif F Q1129, which typically does not have direct contact with NTP. The triphosphate moiety is stabilized by the invariant motif F R1134 and several other basic residues in the NTP entry channel (K662, K729, and K1376). Although the interaction details are quite different between the regular base pairing mode and this alternative mode, the structural elements involved in 1664-TP binding are largely the same. Taken together, these structural observations provide a solid basis for clarifying the unique and effective intervention of 1664-TP.
1664-TP can be slowly incorporated into SARS-CoV-2 and EV71 elongation complexes through Watson-Crick base pairing
Nucleic acid polymerases have different enzymatic properties at different phases of synthesis. The above AMP incorporation assays characterize the very early stage of RNA synthesis. To obtain a comprehensive understanding of 1664-TP intervention, we assembled an elongation complex (EC) using established methods in SARS-CoV-2. By utilizing a T56/P10*3 construct (Fig. 5a, P10*3 denotes three 10-mer molecules that mimic one 30-mer molecule to anneal with the template RNA T56. However, only the P10 annealing at the very downstream can be extended.), one P10 molecule can be extended to yield a 14-mer product (P14) by the addition of a CACA sequence, resulting in a highly stable P14-containing EC with a half-life of more than 70 h at 16 °C19. After the removal of CTP and ATP for EC assembly by anion exchange chromatography, the addition of GTP (100 μM) or GTP/ATP (100 μM each) resulted in efficient conversion of the 14-mer to 16-mer or 19-mer products within 5 min, respectively (Fig. 5b, lanes 3 and 9). When GTP was provided as the only NTP substrate, it induced misincorporation to yield the 17-mer (Fig. 5b, black triangle) and allowed subsequent synthesis (Fig. 5b, lanes 3–7). In both cases, the 20-mer product was likely derived from another misincorporation event (Fig. 5b, lanes 2–13). When GTP and 1664-TP were provided at 100 μM each, the 19-mer product slowly accumulated within the first hour, and the 17-mer intermediate product (Fig. 5b, red triangle, according to 1664-MP incorporation, and MP denotes the NMP form) was clearly evident, suggesting that 1664-TP can lead to slow incorporation in SARS-CoV-2 RdRP EC (Fig. 5b, lanes 14–19). Notably, a small amount of GTP-derived 17-mer was also observed in this reaction. Both the number of GTP- and 1664-TP-derived 17-mers decreased over time, demonstrating that the incorporation of GMP or 1664-MP at this position did not terminate the synthesis. In contrast, when chain-terminating 3′-deoxy-ATP (3′-dATP) and GTP were provided at 100 μM each, the 3′-dATP-derived 17-mer (Fig. 5b, blue triangle) did not diminish over time (Fig. 5b, lanes 20–25), further supporting the non-chain-terminating feature of 1664-TP. Notably, DNA complementary to T56 was used to help resolve the RNA products in these assays as described previously19.
a The T56/P10*3 construct used to assemble a 14-mer-containing SARS-CoV-2 RdRP EC (EC14) that further incorporates 1664-MP in a multi-nucleotide incorporation assay. The solid gray bars indicate the boundaries between P10 molecules; the dashed gray lines indicate the incorporation sites. b 1664-MP can be incorporated by the SARS-CoV-2 RdRP EC and allows subsequent synthesis. A DNA complementary to the T56 RNA (cDNA) was used to resolve the product RNA in the denaturing PAGE. The black, red, and blue triangles indicate the 17-mer derived from GMP misincorporation, 1664-MP incorporation, and 3′-dATP incorporation directed by the same templating uridine, respectively. The underlined sequence directs the synthesis of 15-to-19-mer products. Experiment was repeated twice independently with similar results.
We also tested the incorporation of 1664-MP into ECs formed by enterovirus 71 (EV71) 3Dpol, an RdRP that shares high active site structural similarity with SARS-CoV-2 nsp12 and also utilizes a primer-dependent initiation mechanism19,31,32. When 1664-TP was provided as the only NTP substrate in a single-nucleotide incorporation assay in the preassembled EV71 RdRP EC, the incorporation of 1664-MP was evident, albeit at a much slower rate than that in the presence of ATP when magnesium was provided as the divalent metal ion (Fig. 6a, compare lanes 13–16 with lanes 21–24). When manganese replaced magnesium as the divalent metal ion, the incorporation of 1664-MP occurred much faster (Fig. 6a, compare lanes 21–24 with lanes 25–28). To determine whether HNC-1664 inhibits authentic EV71 infection, we determined an IC50 value of 0.85 μM in Vero E6 cells (Fig. 6b).
a 1664-MP can be incorporated by an EV71 RdRP EC. Left: The T35/P10 construct used to assemble a 16-mer-containing EV71 RdRP EC (EC16) that further incorporates 1664-MP in a single-nucleotide incorporation assay. A DNA complementary to the T35 RNA was used to help release the product RNA from T35 in the denaturing PAGE. Experiment was repeated twice independently with similar results. b Dose-response curve of HNC-1664 for inhibition of EV71. The inhibitory effect of HNC-1664 was calculated relative to the vehicle control group (n = 4). The statistical procedures for collecting and processing the data were performed with GraphPad Prism version 8.0. The IC50 value is presented as the mean ± SD of four independent experiments. c 1664-MP can be incorporated into EV71 RdRP ECs through Watson-Crick base pairing. Left: A 1664-MP-containing EV71 RdRP EC crystal structure with a 2Fo-Fc electron density map (contoured at 1.2σ) overlaid. Base-pairing close contacts (3.5 Å or shorter in distance between non-hydrogen atoms) are indicated by purple dashed lines. Right: A superimposition of the 1664-MP-containing structure and a reference structure (in gray) with a U:A base pair at position -1. The coloring scheme, labeling of RdRP motifs and display of invariant residues are the same as those in Fig. 4g.
To understand how 1664-MP is incorporated into RdRP EC, we solved a 3.0-Å crystal structure of a 1664-MP-containing EV71 RdRP EC (Supplementary Table 3; Supplementary Fig. 8c). The EC was assembled and crystallized through an established method by extending the P10 primer in the T35/P10 construct to a 16-mer product33, and then 1664-TP and manganese ions were added to the EC crystal soaking solution to allow the incorporation of 1664-MP. The structure revealed that 1664-MP was translocated to position -1 (Figs. 6c, -1 is the position immediately upstream of the RdRP active site). The hallmark density of the iodo group confirms the Watson-Crick base pairing mode in this structure (Fig. 6c, left). By comparing the 1664-MP-containing structure with an EV71 RdRP EC structure containing a regular U:A base pair at -1 position, we found that the positions of both the purine base and ribose, the base pair geometry with the templating uridine, and the conformations of key side chains in motifs A/B/C for NTP recognition are highly consistent (Fig. 6c, right). These observations suggest that once incorporated, 1664-MP can mimic AMP well and may readily promote subsequent RNA synthesis.
Discussion
Owing to its indispensable role in viral RNA synthesis, as well as the lack of host homologs in human cells and structural conservation of its active site among RNA viruses, RdRP is an attractive target for antiviral drug development8,34. NA compounds are ideal candidates for developing small-molecule drugs against viral RdRPs. To date, at least three categories of approved viral RdRP-targeting NA drugs have been identified. The first category is represented by the uridine analog sofosbuvir, with key modifications at the ribose 2′-position that exhibits immediate chain-terminating features under most circumstances17. The prominent example in the second category is the adenosine analog remdesivir, with a characteristic cyano group at the ribose 1′-position, which causes unique delayed intervention in certain virus systems, including SARS-CoV-2 and EV7119,20. Notable examples in the third category include favipiravir and molnupiravir, which do not have key modifications in the ribose. With their base substitution (in favipiravir) or modification (in molnupiravir), which results in base pairing ambiguity at both the incorporation and templating stages, these NAs lead to a high frequency of errors in viral genome replication and catastrophe lethality to RNA viruses35,36,37. To be effective, compounds in all three categories first need to be incorporated. In contrast, HNC-1664, which was characterized in this work, can effectively compete with ATP in a non-catalytic manner through an alternative base pairing mode that may be taken by different RNA virus families, thus opening an avenue for developing NA inhibitors with broad-spectrum potential. The modification at the 2′-position of HNC-1664 likely plays a key role in disfavoring the regular NTP binding mode, as the critical 2′-hydroxyl is replaced by hydrogen, while the 2′-C-hydrogen on the other side of the ribose is replaced by fluorine. Consequently, the purine base of 1664-TP has the opportunity to adopt the “flipped” conformation, allowing 1664-TP to establish enough stabilizing interactions at rather tight active sites. The discovery of this “abnormal” NTP binding mode may lead to a revolution in NA drug design if the non-catalytic intervention can indeed be as effective and selective as traditional incorporation-based intervention. Since 1664-TP mainly inhibits RNA synthesis through the non-catalytic intervention, the mutagenic potential common for incorporation-based NAs is less of a concern for 1664-TP. More importantly, the observed “flipped” conformation generates drastically different design spaces when the NTP binding pocket is re-explored. In particular, the ribose moiety resides between the distal part of the ring finger subdomain and the -1 product nucleotide, and no longer faces the relatively tight palm domain pocket near motifs A and B, possibly allowing ribose modifications with large-volume groups.
Compared with most other ribose-modified NAs, HNC-1664 displays much broader-spectrum antiviral effects in vitro against different families of RNA viruses. Intriguingly, HNC-1664 not only shows anti-SARS-CoV-2 delta efficacy in vivo but also that mutations of the viral genome have little effect on the sensitivity of HNC-1664 to VOCs. Notably, in this work, on the basis of the data of the prototype strains, a variety of prevalent emerging and ongoing SARS-CoV-2 VOCs are comprehensively employed to test their sensitivity to the compound. A possible explanation might be that multiple and multifunctional binding modes confer strong and stable binding activity between conserved sites of viral RdRP and 1664-TP. Interestingly, however, the IC50 values of the XBB1.1.18.1, HK.3.1, and BF.7.14 variants are different from that of WIV04. We originally speculated that mutations in nsp12 might be responsible for this phenomenon. We have carefully checked the variants of SARS-CoV-2 used in this study and found that there are no obvious nsp12 mutations in the RdRP active site. The existing nsp12 mutations might not serve as targets of HNC-1664, and the difference in replication kinetics of the WT and SARS-CoV-2 VOCs might truly cause differences in the IC50 values of HNC-1664. The potential cytotoxicity of HNC-1664 is a limitation of our prior study. In fact, we attempted to optimize the structure of HNC-1664. Despite a series of cyclic optimizations, the reborn compounds still exhibit nonnegligible cytotoxicity at marginally high concentrations on certain types of cells. Further studies are warranted to improve the preclinical potential of this compound, and we will consider more strategies for enhancing its inhibitory activity and decreasing its cytotoxicity. Additionally, it is essential to verify whether key binding site mutants confer resistance to HNC-1664 via biochemical and cellular assays, and whether HNC-1664 has inhibitory effects on additional families of RNA viruses via this unusual NTP binding mode.
Taken together, our findings demonstrate that HNC-1664 could be a promising candidate for broad-spectrum antiviral efficacy against different families of RNA viruses. Strikingly, HNC-1664 exhibits two different binding modes with various types of RdRPs, which warrants further development and provides inhibitor resources and structural insights for tackling current or potential future RNA virus outbreaks.
Methods
Cell lines and viruses
Vero-E6, Caco-2, Huh-7, RD, and BHK-21 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 100 U/ml penicillin and 100 μg/ml streptomycin, respectively. The SARS-CoV-2 wild-type strain (WIV04, accession no. EPI_ISL_402124), the alpha variant (B.1.1.7, 1633.06. IVCAS 6.7512), the beta variant (B.1.351, NPRC2.062100001), the delta variant (B.1.617.2; GWHBEBW01000000) and the Omicron variant (B.1.1.529, CCPM-B-V-049-2112-18) were kindly provided by the National Virus Resource Center, Wuhan Institute of Virology, Chinese Academy of Sciences. The assays for the abovementioned authentic SARS-CoV-2 prototype and prevalent emerging strains in this study were performed at the biosafety level 3 (BSL-3) facility of the Wuhan Institute of Virology, Chinese Academy of Sciences. The assays for SARS-CoV-2 XBB.1.18.1 (GenBank: OQ653373.1), HK.3.1 (GenBank: OR647475.1), and BF.7.14 (GenBank: OQ653373.1) variants used in this study were performed at the biosafety Level 3 (BSL-3) facility in the Division of Life Sciences and Medicine, University of Science and Technology of China. The LCMV Armstrong strain was rescued as described previously13. Briefly, the generations of pT7-LCMV-S-P2A-eGFP and pT7-LCMV-L were based on the Armstrong strain ARM53b S segment (GenBank: AY847350.1) and L segment (GenBank: AY847351.1), respectively. BSR-T7 cells were seeded in 6-well plates at a density of 2 × 106 cells per well. In accordance with the manufacturer’s instructions for Lipo3000 transfection reagent, BSR-T7 cells were transiently transfected with 300 ng of pCAGGS NP, 600 ng of pCAGGS L, 300 ng of pT7-LCMV-S-P2A-eGFP, and 600 ng of pT7-LCMV-L. The successful rescue was directly monitored via fluorescence microscopy (eGFP), and the supernatant was collected at 72 h post-transfection, followed by propagation and titration in BSR-T7 cells.
Antiviral effects of HNC-1664 in vitro
Vero E6 and/or Caco-2 cells were seeded in 96-well plates at a density of 1 × 104 cells per well and incubated for 16 h at 37 °C. The cells were treated with HNC-1664 or remdesivir and infected with the SARS-CoV-2 wild-type strain or its variants, which included alpha, beta, delta, and Omicron strains, at a multiplicity of infection (MOI) of 0.5 and with XBB.1.1, HK.3.1, and BF.7.14 at an MOI of 0.1. The cells were collected at 24 h post-infection, and the quantification of NP expression was evaluated via IFA. The quantification of NP expression was used to calculate the inhibition of infection relative to the vehicle control group, and the IC50 values were generated from inhibition curves. Three independent experiments were conducted, each with three technical replicates. The antiviral effects of HNC-1664 against HCoV-OC43 and HCoV-229E were determined in RD cells and Huh-7 cells, respectively. The cells were infected with HCoV-OC43 and/or HCoV-229E at an MOI of 0.5. At 24 h.p.i., viral RNA was extracted from the total cells, and viral gene expression was analyzed by RT-qPCR to determine the IC50 values relative to the vehicle control group. Three independent experiments were conducted, each with two technical replicates. To determine the antiviral effect of HNC-1664 against LCMV, Huh-7 cells were infected with the LCMV Armstrong strain at an MOI of 0.5. Infected cells were collected at 24 h post-infection, and the data were analyzed via IFA. The inhibition of infection was calculated relative to the vehicle control group, and the IC50 value was generated from an inhibition curve. Three independent experiments were conducted, each with three technical replicates. To determine the antiviral effect of HNC-1664 against EV71, Vero E6 cells were seeded in 24-well plates at a density of 1 × 105 cells per well and incubated for 16 h at 37 °C. Then, the cells were treated with HNC-1664 and infected with the EV-A71 strain at an MOI of 0.1. Infected cells were collected at 24 h post-infection, and viral gene expression was analyzed via RT-qPCR to determine the IC50 values relative to the vehicle control group. Four independent experiments were conducted, each with two technical replicates. All of the quantification data are presented as the mean ± SD.
IFA assay
The cells were fixed with 4% paraformaldehyde and permeabilized with phosphate-buffered saline (PBS) containing 0.2% Triton X-100 for 15 min. The cells were subsequently blocked with 5% fetal bovine serum (FBS; Gibco), followed by treatment with a primary antibody against the SARS-CoV-2 N protein rabbit mAb (ABclonal; A20021) at a 1:1000 dilution. After six rinses with PBS, the cells were stained with the secondary antibody DyLight 488-labeled anti-rabbit IgG (KPL, Gaithersburg, MD, USA). Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI) according to the manufacturer’s instructions (Sigma-Aldrich, USA). The cells were imaged using an Operetta high-content imaging system (PerkinElmer), and the number of infected cells and DAPI-positive cells per well were calculated using the associated Harmony 3.5 software.
Cell viability assay
Vero E6, Caco-2, HEK293T, RD, and/or Huh-7 cells were seeded in 96-well plates at a density of 1 × 104 cells per well and incubated for 16 h at 37 °C. The cells were treated with HNC-1664 at concentrations ranging from 0.00001 μM to 100 μM or with the vehicle. For the cytotoxic effects of HNC-1664 on transfected Vero E6 and HEK293T cells, treatment with HNC-1664 was performed 4 h post-transfection. After 24 h of HNC-1664 incubation, the effect of HNC-1664 on cell viability was evaluated via a CCK-8 cell counting kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The absorbance was measured via an EnSpire multimode plate reader at an emission wavelength of 450 nm. The cell viability was calculated relative to the vehicle control group. At least four independent experiments were conducted, each with three technical replicates. All of the quantification data are presented as the mean ± SD.
Minigenome assay
The minigenomes of LASV and LCMV were generated by replacing the S segment ORFs with ZsGreen (ZsG) and Gaussia princeps luciferase (gLuc)38. To evaluate the inhibitory effect of HNC-1664 on the LASV and/or LCMV minigenome, cells were transfected with DNA according to the manufacturer’s instructions of Lipo3000 transfection reagent. Briefly, vero E6 and/or HEK293T cells were seeded in 96-well plates at a density of 1 × 104 cells per well and incubated for 16 h at 37 °C. For the LASV minigenome assay, Vero E6 cells were transfected with 30 ng of pLASV-S mini, 50 ng of pCAGGS-LASV NP, and together with 10 ng of pCAGGS-LASV L per well. For the LCMV minigenome assay, HEK293T cells were transfected with 25 ng of pLCMV-S mini, 20 ng of pCAGGS-LCMV NP, and together with 10 ng of pCAGGS-LCMV L per well. The empty vector plasmid (pCAGGS) was cotransfected with NP and the S mini expression plasmid as a negative control. At 4 h post-transfection, HNC-1664 or ribavirin was added at the indicated concentrations. After 24 h of incubation, the luciferase activity of the cell lysates was evaluated relative to the vehicle control group. Five independent experiments were conducted, each with three technical replicates for LASV mini and four technical replicates for LCMV mini. All quantification data are presented as the mean ± SD.
Antiviral activity of HNC-1664 in vivo
Female K18-hACE2 transgenic mice aged 5–6 weeks were obtained from Gempharmatech Co., Ltd. (Nanjing, China). The use of K18-hACE2 transgenic mice according to the standards for the use and care of laboratory animals was approved by the Institutional Review Board of the Wuhan Institute of Virology, Chinese Academy of Sciences. The in vivo study was approved by Wuhan Institute of Virology, Chinese Academy of Sciences (WIVA25202110). All animals were randomly assigned to three groups and intranasally inoculated with either 1 × 105 PFU of the SARS-CoV-2 strain delta, which was prediluted in 20 μl of MEM. At 2 and 14 h.p.i., the mice were administered oral gavage 50 mg kg−1 HNC-1664 diluted in vehicle solution (5% DMSO, 5% Solutol HS-15, and 90% normal saline) or 500 mg kg−1 molnupiravir diluted in normal saline or vehicle solution only as a control group. For therapeutic treatment, the mice were subsequently administered HNC-1664, molnupiravir or vehicle solution by oral gavage twice daily beginning at 1-day post infection (d.p.i.). Converting on the basis of body surface area, the dosage used for the treatment of human patients was 50 mg kg−1 dose (mouse) × 0.08 (conversion factor) = 4 mg kg−1 dose, which would mean that a 60 kg person requires a 4 mg kg−1 dose × 60 kg × 2 dose = 480 mg per day. The mice were sacrificed at 4 d.p.i., and lung tissues were sampled for immunohistochemical and hematoxylin-eosin (H&E) staining. The nasal turbinate, trachea, and lungs were collected and ground in 1 mL of DMEM. For the determination of viral titers per gram of tissue, the supernatant samples were collected after centrifugation and assessed via plaque assay. Moreover, the number of viral copies per gram of tissue was determined via RT-qPCR.
Hematoxylin & eosin (H&E) and immunohistochemical staining
In brief, the lung tissues of infected mice were fixed in 10% formalin for more than 7 days. The fixed samples were then embedded in paraffin wax and sectioned via a TP1020 Leica semienclosed desktop tissue processor. The slices were dried and fixed on slides overnight at 37 °C. Then, for dewaxing, the slices were successively washed in environmentally friendly dewaxing solution and anhydrous ethanol three times. For hematoxylin and eosin staining, the slices were stained with hematoxylin and eosin. For the immunohistochemical staining assays, the slices were treated with 3% hydrogen peroxide at room temperature (r.t.) and shielded from light for 25 min to block endogenous peroxidase, followed by incubation with 3% BSA for 30 min. Rabbit anti-SARS-CoV-2-NP immune serum (stored at Servicebio Co. Ltd., 1:100) and an HRP-labeled goat anti-mouse IgG antibody (GB23301, Servicebio, 1:200) were used as the primary and secondary antibodies, respectively. Signal detection was performed with a 3,3’-diaminobenzidine (DAB) substrate kit (G1212, Servicebio). The nucleus was labeled with hematoxylin. Images were taken under a white light microscope.
Plaque assay
To determine viral titers in mice, nasal turbinate, trachea, and lungs were harvested and ground in 1 mL of DMEM at 4 h.p.i. Vero E6 cells were seeded in 24-well plates at a density of 1 × 105 cells per well and incubated for 16 h at 37 °C. The supernatants of the collected tissues were diluted 10-fold and added to a 24-well culture plate. After infection at 37 °C for 1 h, the supernatant was discarded, and the cells were overlaid with medium containing 1% methylcellulose and incubated for 4 days at 37 °C. The viral titers were subsequently determined via crystal violet staining.
Evaluation of viral quantity by RT-qPCR
Lysates of HCoV-OC43- and HCoV-229E-infected cells were extracted via TRIzol reagent. The quantification of viral RNA was performed with the primers OC43 M-F (5′-ATGTTAGGCCGATAATTGAGGACTAT-3′), OC43 M-R (5′-AATGTAAAGATGGCCGCGTATT-3′), 229E M-F (5′-TTCCGACGTGCTCGAACTTT-3′) and 229E M-R (5′-CCAACACGGTTGTGACAGTGA-3′). The level of GAPDH RNA was calculated with the primers Human GAPDH-F (5′-GAAGGTGAAGGTCGGAGTC-3′) and GAPDH-R (5′-GAAGATGGTGATGGGATTTC-3′) as controls. The quantification of EV71 RNA copies was performed with the primers EV71-F (5′-GGCCATTTATGTGGGTAACTTTAGA-3′) and EV71-R (5′-CGGGCAATCGTGTCACAAC-3′). GAPDH RNA level was calculated with the primers Vero E6 GAPDH-F (5′-TCCTTGGAGGCCATGTGGGCCAT-3′) and GAPDH-R (5′-TGATGACATCAAGAAGGTGGTGAAG-3′) as controls. To determine the viral RNA loads in the mice, viral RNA was extracted from nasal turbinate, trachea, and lungs via a Virus DNA/RNA Extraction Kit 2.0 (Vazyme, Nanjing, China). Then, following the instructions of the HiScript® II One Step qRT-PCR SYBR® Green Kit (Vazyme, Nanjing, China), the quantification of viral RNA was performed with the following primers: ORF1a/b-F (5′-CCCTGTGGGTTTTACACTTAA-3′) and ORF1a/b-R (5′-ACGATTGTGCATCAGCTGA-3’). Serial dilutions of the SARS-CoV-2 ORF1a/b gene control plasmid were tested to generate standard curves.
Preparation of the viral RdRP or RdRP complex
For the structural and pre-elongation enzymology characterization of SARS-CoV-2 RdRP, plasmids containing SARS-CoV-2 nsp12, nsp8, and nsp7 were transformed into E. coli Rosetta (DE3). A single colony was added to Luria-Bertani medium supplemented with 50 μg/ml carbenicillin at 37 °C until the optical density reached 0.7, and the growth temperature was subsequently reduced to 16 °C. Afterward, 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added to induce expression for 20 h. Afterwards, the cell pellets were harvested and resuspended in lysis buffer (50 mM Tris-HCl (pH 8.0), 500 mM NaCl, 4 mM MgCl2, and 5% glycerol) supplemented with 0.5 mM phenylmethylsulfonyl fluoride (PMSF) (Biosharp) and lysed by an ultrasonic homogenizer (SCIENTZ). Nickle column affinity and size exclusion chromatography of nsp12, nsp8 and nsp7 were performed according to previous protocols19. To characterize the elongation phase of the SARS-CoV-2 RdRP used for enzymology, nsp12, nsp8, and nsp7 were prepared as previously described19,39. To determine the inhibitory mechanism of 1664-TP, a 1.5 mL reaction mixture of 10 μM nsp12, 10 μM nsp7, 20 μM nsp8 and 10 μM RNA was mixed in reaction buffer (50 mM HEPES (pH 7.2), 100 mM NaCl, 5 mM MgCl2 and 4 mM DTT) with r.t. for 1 h before being loaded into the superdex200 increase column pre-equilibrated with gel filtration buffer (50 mM HEPES (pH 7.2), 100 mM NaCl, 5 mM MgCl2 and 4 mM DTT. The peak fractions were collected, concentrated to 5.0 mg/mL and incubated with 1 mM 1664-TP on ice prior to cryogenic specimen preparation.
For LASV L protein production, the pPICZ-B vector-based yeast expression construct (kindly provided by Dr. Zengqin Deng) expressing the Lassa mammarenavirus (Josiah strain) L protein fused with a C-terminal PreScission protease cleavage site and a GFP-deca-histidine (GFP-His*10) tag was transformed into the yeast P. pastoris (SMD1163H) for protein production. For protein purification, yeast cells were harvested and resuspended in lysis buffer containing 50 mM NaH2PO4 (pH 8.0), 0.5 M NaCl, 10 mM imidazole, 10% (vol./vol.) glycerol, 4 mM MgCl2, and 1 mM PMSF. The cells were lysed by an AH-2010 homogenizer at 14,500 psi (ATS Engineering) and subsequently centrifuged at 17,000 rpm in an F21-8 × 50 y rotor (Thermo Fisher Scientific) for 60 min at 4 °C. The supernatant was collected, loaded onto Ni2+-charged resin FF and washed with a series of wash buffers containing 50 mM NaH2PO4 (pH 8.0), 500 mM NaCl, 20/40/60 mM imidazole, and 10% (vol./vol.) glycerol. The L-GFP fusion protein was eluted with wash buffer supplemented with 300 mM imidazole and subsequently digested with PreScission protease at 4 °C overnight to remove the C-terminal GFP-His*10 tag. Further purification was performed via a HiTrap Heparin HP column (GE Healthcare) followed by a Superose 6 Increase 10/300 Gl size exclusion column (GE Healthcare) equilibrated with 20 mM HEPES (pH 7.5), 500 mM NaCl, 5 mM tris-(2-carboxyethyl) phosphine (TCEP) (pH 7.5), and 10% (vol./vol.) glycerol. The purified L protein was pooled and concentrated to 30 mg/mL, flash-frozen in liquid nitrogen as 2–10 μl aliquots, and stored at −80 °C for single use.
For EV71 3Dpol protein production, the pET26b-Ub vector-based plasmid containing the EV71 (strain SK-EV006-LPS1) 3Dpol (RdRP) gene with a C291M mutation was used40. Cell growth, IPTG induction, cell harvesting, cell lysis, protein purification, and protein storage were performed according to previously described methods with minor modifications22,41. Briefly, the cells were grown at 37 °C in LB medium with 25 μg/mL kanamycin and 20 μg/mL chloramphenicol until the optical density at 600 nm (OD600) reached 0.6, after which they were cooled to r.t. IPTG was added to a final concentration of 0.5 mM, and cells were subsequently grown at 25 °C for 11–12 h until they reached an OD600 of 1.3–1.4 before harvesting. Three successive chromatography steps were performed using HisTrap HP (Nickel affinity), HiTrap Q HP (anion exchange), and HiLoad 16/600 Superdex 200 pg (gel filtration) columns (GE Healthcare). For the anion-exchange step, the column was equilibrated with a buffer containing 113 mM NaCl, 25 mM Tris (pH 8.5), 0.1 mM EDTA, and 20% (vol./vol.) glycerol, and the protein was eluted via a linear gradient to 620 mM NaCl. For the gel filtration step, the column was equilibrated with 300 mM NaCl, 5 mM Tris (pH 7.5), 20% (vol./vol.) glycerol. NaN3 was not supplied in the lysis buffer and chromatography buffers.
RNA preparation
The RNA templates were either prepared through a T7 RNA polymerase-glmS ribozyme-based method (T56, T35, T32)42 with denaturing polyacrylamide gel electrophoresis (PAGE) purification and electro-elution replacing the affinity purification step43, or chemically synthesized (T30) (Integrated DNA Technologies). The RNA primer P10 and vRNA 0–13 were chemically synthesized (Integrated DNA Technologies or Sangon Biotech). T30 was synthesized by an in-house H-8 synthesizer (K&A Laboratories). P10 was annealed to T35 or T56 at a 1.1:1 or 3.1:1 molar ratio, respectively.
Viral RdRP activity assays
The RNA oligonucleotides used for the pre-elongation SARS-CoV-2 polymerase assay were purchased from Accurate Biology. The sequence used for the replication assay was FAM-UUUUUCAUGCUACGCGUAGUUUUCUACGCGUAGCAUG. The sequence of the RNA duplex was modified from Hillen et al., and the duplex contained a primer (5′-CUACGCGUAGCAUGG-3′) and a template (5′-UUUUUCUAUGCUACGCGUAG-3′)44. The RNA was annealed in annealing buffer containing 10 mM HEPES-Na (pH 7.5) and 50 mM NaCl by heating the mixture to 90 °C for 3 min and then gradually cooled to r.t. The 20 μL reactions included RNA (1 μM), nsp12 (1 μM), nsp8 (2 μM), and nsp7 (1 μM) in 50 mM Na-HEPES (pH 7.0), 50 mM NaCl, 5% (v/v) glycerol, 5 mM MgCl2 and 4 mM DTT. The incorporation assay was initiated by the addition of 250 μM ATP at 30 °C and stopped by the addition of 2× stop buffer (10 M urea, 1× TBE, 0.02% (wt./vol.) xylene cyanol, and 0.02% (wt./vol.) Bromphenol blue and separated on 8 cm × 8 cm 20% acrylamide gels in 1× TBE buffer supplemented with 7 M urea. 5-FAM-labeled RNA products were visualized by a Typhoon 7000 FLA Imager (GE Healthcare Life Sciences).
For the LASV RdRP single-nucleotide incorporation assay, a 10-μl reaction mixture containing 6 μM L protein, 4 μM RNA T30/P3, 0.25 mM ATP only, or 0.25 mM ATP with 0.05/0.1/1 mM 1664-TP or 0.25 mM 1664-TP only in reaction buffer (5 mM MES (pH 6.5), 5 mM MgCl2, 5 mM TCEP) was incubated at 25 °C for 10, 30, or 60 min. The reaction was stopped by adding an equal volume of stop solution (95% (vol./vol.) formamide, 20 mM EDTA (pH 8.0), 0.02% (wt./vol.) xylene cyanol).
For the multi-nucleotide incorporation assay using SARS-CoV-2 RdRP, the T56-derived P14-containing EC (EC14) was assembled and purified as described previously19. A typical 20 μL reaction mixture containing 100 μM GTP, 100 μM 1664-TP or ATP, and 4 μM EC14 in a reaction buffer of 50 mM HEPES (pH 7.0), 100 mM NaCl, 4 mM MgCl2, and 4 mM DTT was incubated at 25 °C. After various reaction durations (0-120 min), the reaction was stopped by adding an equal volume of stop solution.
For the single-nucleotide incorporation assay using EV71 RdRP, the T35-derived P16-containing EC (EC16) was assembled and purified as described previously40. A typical 20-μL reaction mixture containing 300 μM ATP or 300 μM 1664-TP, and 4 μM EC16 in a reaction buffer of 50 mM HEPES (pH 7.0), 30 mM NaCl, 50 mM KCl, 5 mM MgCl2, and 5 mM TCEP was incubated at 22.5 °C. After various reaction durations (2–120 min), the reaction was stopped by adding an equal volume of stop solution. The RNA species were resolved via 20% polyacrylamide-7 M urea gel electrophoresis until bromphenol blue migrated to approximately three-fourths of the vertical gel dimension and then visualized by staining with Stains-All (Sigma-Aldrich).
Cryo-EM sample preparation of SARS-CoV-2 RdRP in complex with 1664-TP
In total, 3 μL of protein-RNA complex solution at 5 mg/mL (with 0.003% OG) was added to a H2/O2 glow-discharged, 200-mesh Quantifoil R2/1 copper grid (Quantifoil, Micro Tools GmbH, Germany). The grid was then blotted for 3.5 s, incubated for 5 s at a blot force of 0 at 4 °C and 100% humidity and then frozen in liquid ethane via a Vitrobot (Marker IV, Thermo Fisher Scientific, USA). Cryo-EM data were collected with a 300 keV Titan Krios electron microscope (Thermo Fisher Scientific, USA) and a K3 Summit direct electron detector (Gatan, USA). Images were recorded via EFTEM at 105,000× magnification with a calibrated pixel size of 0.82 Å/pixel. The exposure time was set to 3 s, with a dose rate of 17.8 electrons/second/Å2. All images were automatically recorded via EPU software (Thermo Fisher Scientific, USA), and a total of 3,705 movie stacks were collected with a defocus range from −1.6 µm to −3.2 µm. The data processing procedure was carried out via standard pipelines in cryoSPARC. All dose-fractioned movies were processed in a series of steps, including motion correction, CTF estimation, particle picking and extraction, 2D classification, ab initio 3D reconstruction, heterogeneous 3D refinement and nonuniform homogeneous refinement.
Model construction and refinement of the SARS-CoV-2 RdRP complex
Coordinates for nsp7, nsp8, nsp12, and RNA were obtained from PDB entry 6YYT. These coordinates were rigidly fitted into the cryo-EM map using UCSF Chimera45 by applying the “fit in map” tool. This provided an initial placement of the structure into the density map while maintaining fidelity to the input coordinates. Following the initial rigid fitting, manual inspection in Coot46 was performed to refine regions where automatic fitting did not achieve sufficient accuracy, such as areas with weaker density or ambiguous structural features. Manual adjustments were performed iteratively, with the density level set at 3.33 rmsd to ensure the density was well-visualized without obscuring finer details. Manual adjustments were deemed sufficient when the following criteria were met: (1) Fit to density: Backbone and side-chain atoms showed a close fit to the cryo-EM density map contours. At this stage, any residual unmodeled density in these regions was negligible, indicating that no further refinement was necessary. (2) Geometrical validation: Each round of manual adjustments was followed by real-time geometrical validation using Coot’s built-in tools (Ramachandran plot analysis, rotamer checks). Adjustments were repeated until no steric clashes or outliers were detected, and the model adhered to protein geometry standards. (3) Convergence of refinement: In areas of weaker density, refinement using phenix.real_space_refine47 was employed. Manual examination was deemed sufficient once automated refinement no longer resulted in significant changes in atom positions. Refinement was carried out using phenix.real_space_refine with several parameter sets, including: minimization_global for global geometry minimization, adp (atomic displacement parameters) with an isotropic B-factor refinement strategy, ensuring the B-factors corresponded to the local resolution of the map, and secondary structure restraints applied to ensure that α-helices and β-sheets retained their correct geometry.
For the compound HNC-1664, only the chemical structure in the form of a SMILES string was available. Therefore, the electronic Ligand Builder and Optimization Workbench (eLBOW), a module of PHENIX, was used to generate its coordinate and CIF file for refinement. Docking of the compound into the cryo-EM map was carried out using Chimera (command: fit #model #map search 10). The top-scoring docking models were manually examined in Coot, focusing on fitting the ligand into well-defined density regions and avoiding clashes with nsp12 residues. Manual adjustments were primarily limited to side chains interacting with HNC-1664, using real-space refinement tools in Coot, with the density level set at 3.33 rmsd for ligand positioning. The final model, consisting of nsp7, nsp8, nsp12, RNA, and HNC-1664, was refined using phenix.real_space_refine with the same parameter set as described previously. The final model was evaluated using MolProbity, and the statistics of the map reconstruction and model building are summarized in SI Table 1. Figures were generated using Chimera and ChimeraX48.
Cryo-EM sample preparation of LASV RdRP EC
To prepare the Cryo-EM samples, a 3.5 μl reaction mixture containing 6 μM LASV L protein, 8 μM vRNA 0-13 (the 5′-terminal 13-nucleotide region of the L segment with an additional guanosine nucleotide at the 5′-end), 8 μM template RNA T32/pGGA (5′-GGGAGAUGAAAGUCUCCAACUGAAGAGUCCAA-3′), 0.3 mM ATP in reaction buffer (50 mM HEPES (pH 7.0), 5 mM MgCl2, and 5 mM TCEP) was incubated at 25 °C for 30 min. 1664-TP was added to the reaction at a final concentration of 500 μM and incubated for 60 min on ice before sample preparation. Aliquots of 3.5 μl of the reaction mixture were immediately loaded onto glow-discharged holy carbon copper grids (Quantifoil, R1.2/1.3). The grids were blotted for 3 s at 100% humidity, 16 °C and a blot force of 15 and then plunged frozen in liquid ethane at liquid nitrogen temperature using a Vitrobot Mark IV (Thermo Fisher Scientific). The ø 55/20 mm blotting paper (GE Healthcare) was used for plunge freezing. The grids were stored in liquid nitrogen before data collection.
Cryo-EM data collection, processing and structure determination of LASV EC
Cryo-EM data were collected with a 300 keV CryoARM300 electron microscope (JEOL, Japan) and a K3 direct electron detector (Gatan, USA). Images were recorded at 50k magnification with a calibrated super-resolution pixel size of 0.475 Å/pixel. The exposure time was set to 1.6 s with a total accumulated dose of 40 electrons/Å2. All the images were automatically recorded via serial EM49 with a defocus range from 1.0 µm to 2.0 µm. The statistics for the data collection and refinement are provided in SI Table 2.
The data processing procedure was carried out via standard pipelines in cryoSPARC50. The raw movies were processed in a series of steps, including motion correction, CTF estimation, particle picking and extraction, 2D classification, ab initio 3D reconstruction, heterogeneous 3D refinement and non-uniform homogeneous refinement.
The initial model for the EC structure was generated from the previously reported cryo-EM structure of a LASV RdRP EC structure (PDB 7OJN)51. The initial models were placed and rigid-body fitted into the cryo-EM map via UCSF Chimera45. The model was manually built in Coot46 with the guidance of the cryo-EM map and in combination with real space refinement via PHENIX (Supplementary Fig. 7)47. The data validation statistics are shown in SI Table 2. Although MgCl2 was present in the sample buffer, we did not find a suggestive EM density of catalytic divalent metal ions. Therefore, Mg2+ was not included in the structural model.
EV71 RdRP EC crystallization and 1664-TP soaking
T35-derived P16-containing EV71 RdRP EC crystals were grown by hanging-drop vapor diffusion at 16 °C using a 14 mg/mL EC sample and EC crystal microseeds. Crystals were grown for 2 days in a precipitant/well solution containing 0.17 M ammonium sulfate, 0.2125 M MES (pH 6.5), and 26% (wt./vol.) PEG5000 monomethyl ether, and 15% (vol./vol.) glycerol. 1664-TP soaking trials were performed using EC crystals in a precipitant solution containing 1.4 mM 1664-TP and 10 mM MnCl2 for 24 h.
Crystallographic data processing and structure determination of EV71 EC
The X-ray diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL10U2 at 100 Kelvin (wavelengths: 0.9792 Å). 180° data were typically collected in 0.2° oscillation angle. Reflections were indexed, integrated, merged, and scaled by XDS52. The initial structural solution was obtained via the Phaser molecular replacement program with an EV71 EC structure (PDB entry: 5F8G) as the search model22,53. Manual model rebuilding was performed via Coot, and the model was refined via PHENIX46,54. A 3500-K composite simulated-annealing (SA) omit 2Fo-Fc electron density map was generated via a Crystallography & NMR system (CNS)55. The X-ray diffraction data collection and structure refinement statistics are shown in SI Table 3. Although MnCl2 was present in the sample buffer, we did not find a suggestive electron density for the catalytic divalent metal ions. Therefore, Mn2+ was not included in the structural model.
Statistical analysis
All the statistical analysis and corresponding graphs were generated with GraphPad Prism version 8.0. The data are presented as the mean ± SD. One-way ANOVA (analysis of variance) with Dunnett’s post-hoc test was used to determine statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns no significance.).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All source data for evaluating the conclusions of this study are provided in the Supplementary Information or Source Data files. The cryo-EM maps and atomic coordinates generated in this study have been deposited in the Protein Data Bank (PDB) and the Electron Microscopy Data Bank (EMDB) under accession codes as follows: SARS-CoV-2 RdRP with 1664-TP (PDB 8XKO, EMD-38422); LASV L with 1664-TP (PDB 8XPO, EMD-38557). The crystallographic structure factors and atomic coordinates of the 1664-MP-incorporated EV71 RdRP EC structure have been deposited in the PDB with the accession code 8XPP. Source data are provided with this paper.
References
Reperant, L. A. & Osterhaus, A. D. M. E. AIDS, Avian flu, SARS, MERS, Ebola, Zika. what next? Vaccine 35, 4470–4474 (2017).
Drosten, C. et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 (2003).
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. & Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Boyton, R. J. & Altmann, D. M. Imprinted hybrid immunity against XBB reinfection. Lancet Infect. Dis. 23, 764–765 (2023).
Bobrovitz, N. et al. Protective effectiveness of previous SARS-CoV-2 infection and hybrid immunity against the omicron variant and severe disease: a systematic review and meta-regression. Lancet Infect. Dis. 23, 556–567 (2023).
Yin, Y. & Wunderink, R. G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 23, 130–137 (2018).
Vicenti, I., Zazzi, M. & Saladini, F. SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19. Expert Opin. Ther. Pat. 31, 325–337 (2021).
Moreno, H. et al. A novel circulating tamiami mammarenavirus shows potential for zoonotic spillover. PLoS Negl. Trop. Dis. 14, e0009004 (2020).
Garry, R. F. Lassa fever—the road ahead. Nat. Rev. Microbiol. 21, 87–96 (2022).
Bonthius, D. J. Lymphocytic choriomeningitis virus: an underrecognized cause of neurologic disease in the fetus, child, and adult. Semin. Pediatr. Neurol. 19, 89–95 (2012).
Zapata, J. C. et al. Lymphocytic choriomeningitis virus (LCMV) infection of macaques: a model for Lassa fever. Antivir. Res. 92, 125–138 (2011).
te Velthuis, A. J. Common and unique features of viral RNA-dependent polymerases. Cell Mol. Life Sci. 71, 4403–4420 (2014).
Jia, H. & Gong, P. A structure-function diversity survey of the RNA-dependent RNA polymerases from the positive-strand RNA viruses. Front. Microbiol. 10, 1945 (2019).
Seley-Radtke, K. L., Thames, J. E. & Waters, C. D. 3rd. Broad spectrum antiviral nucleosides-our best hope for the future. Annu. Rep. Med. Chem. 57, 109–132 (2021).
Gane, E. J. et al. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N. Engl. J. Med. 368, 34–44 (2013).
Appleby, T. C. et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 347, 771–775 (2015).
Malone, B. F. et al. Structural basis for substrate selection by the SARS-CoV-2 replicase. Nature 614, 781–787 (2023).
Wu, J. et al. Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex. Cell Rep. 37, 109882 (2021).
Gordon, C. J. et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 295, 6785–6797 (2020).
Gong, P. & Peersen, O. B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl Acad. Sci. USA 107, 22505–22510 (2010).
Shu, B. & Gong, P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl Acad. Sci. 113, E4005–E4014 (2016).
Gong, P. Structural basis of viral RNA-dependent RNA polymerase nucleotide addition cycle in picornaviruses. Enzymes 49, 215–233 (2021).
Furuta, Y. et al. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 100, 446–454 (2013).
Jayk Bernal, A. et al. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N. Engl. J. Med., https://doi.org/10.1056/NEJMoa2116044 (2021).
Sanderson, T. et al. A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes. Nature 623, 594–600 (2023).
Waters, M. D., Warren, S., Hughes, C., Lewis, P. & Zhang, F. Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: the special case of molnupiravir. Environ. Mol. Mutagen. 63, 37–63 (2022).
Lin, C. et al. Design, synthesis, and biological evaluation of novel 7-deazapurine nucleoside derivatives as potential anti-dengue virus agents. Antivir. Res. 149, 95–105 (2018).
Uengwetwanit, T., Chutiwitoonchai, N., Wichapong, K. & Karoonuthaisiri, N. Identification of novel SARS-CoV-2 RNA dependent RNA polymerase (RdRp) inhibitors: From in silico screening to experimentally validated inhibitory activity. Comput. Struct. Biotechnol. J. 20, 882–890 (2022).
Gao, Y. et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368, 779–782 (2020).
Wang, Q. et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell 182, 417–428.e413 (2020).
Campagnola, G., Govindarajan, V., Pelletier, A., Canard, B. & Peersen, O. B. The SARS-CoV nsp12 polymerase active site is tuned for large-genome replication. J. Virol. 96, e00671–00622 (2022).
Gong, P., Kortus, M. G., Nix, J. C., Davis, R. E. & Peersen, O. B. Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts. PloS ONE 8, e60272 (2013).
Tian, L. et al. RNA-dependent RNA polymerase (RdRp) inhibitors: the current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 213, 113201 (2021).
Crotty, S., Cameron, C. E. & Andino, R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl Acad. Sci. USA 98, 6895–6900 (2001).
Jin, Z., Smith, L. K., Rajwanshi, V. K., Kim, B. & Deval, J. The ambiguous base-pairing and high substrate efficiency of T-705 (Favipiravir) Ribofuranosyl 5’-triphosphate towards influenza A virus polymerase. PloS ONE 8, e68347 (2013).
Kabinger, F. et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 28, 740–746 (2021).
Lan, X. et al. Screening and identification of Lassa virus endonuclease-targeting inhibitors from a fragment-based drug discovery library. Antivir. Res. 197, 105230 (2022).
Liu, Q., Wu, J. & Gong, P. Assessment of nucleotide/nucleoside analog intervention in primer-dependent viral RNA-dependent RNA polymerases. STAR Protoc. 3, 101468 (2022).
Wang, M. et al. Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures. Nat. Commun. 11, 2605 (2020).
Tellinghuisen, T. L., Lu, G. & Gong, P. Crystal structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 9, e1003549 (2013).
Batey, R. T. & Kieft, J. S. Improved native affinity purification of RNA. Rna 13, 1384–1389 (2007).
Wu, J. et al. Perturbation in the conserved methyltransferase-polymerase interface of flavivirus NS5 differentially affects polymerase initiation and elongation. J. Virol. 89, 249–261 (2015).
Hillen, H. S. et al. Structure of replicating SARS-CoV-2 polymerase. Nature 584, 154–156 (2020).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D. Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. Sect. D. Struct. Biol. 74, 814–840 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Kouba, T. et al. Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity. Nat. Commun. 12, 7018 (2021).
Kabsch, W. Xds. Acta Crystallogr. Sect. D., Biol. Crystallogr. 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D. Biol. Crystallogr. 66, 213–221 (2010).
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D. Biol. Crystallogr. 54, 905–921 (1998).
Acknowledgements
We thank Dr. Xi Zhou for providing authentic EV71 for determining the IC50 value. We thank Dr. Zihe Rao for providing the plasmids for SARS-CoV-2 RdRP complex production, Dr. Zengqin Deng for providing the plasmid for LASV L protein production, Dr. Jiqin Wu for laboratory assistance and helpful discussions, Dr. Xinyu Wang for preparation of the T30 RNA, and Dr. Bo Shu for helpful discussion in EM data processing. We thank Yongxiang Gao and Peiping Tang from the integrative imaging center of the University of Science and Technology of China for the assistance with Cryo-EM grids screening and data collection. We thank the Center for Instrumental Analysis and Metrology and Core Facility and Technical Support of Wuhan Institute of Virology for providing technical assistance. This study was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB0490000 to S.C., P.G., K.Z., W.W. and J.Z.), the National Key Research and Development Program of China (2022YFC2303300 to P.G., 2022YFC2303700 to K.Z. and S.L., and 2022YFA1302700 to K.Z.), the Guangzhou National Laboratory Major Project (GZNL2024A01010 to P.G.), the National Natural Science Foundation of China (U23A20147 and 3231101030 to P.G., 32371345 to K.Z. and 32301044 to S.L.), the Research Grants Council Collaborative Research Fund of Hong Kong, China (C6036-21G to P.G.), the Creative Research Group Program of the Natural Science Foundation of Hubei Province, China (2022CFA021 to P.G.), the Anhui Provincial Natural Science Foundation (2308085QC80 to S.L.), the Center for Advanced Interdisciplinary Science and Biomedicine of IHM (QYPY20220019 to K.Z.), the Fundamental Research Funds for the Central Universities (WK9100000032 to S.L.), the Key Biosafety Science and Technology Program of Hubei Jiangxia Laboratory, China (JXBS001 to P.G.), and the Knowledge Innovation Program of Wuhan-Shuguang Project, China (2022020801020153 and 2023020201020289 to R.L.).
Author information
Authors and Affiliations
Contributions
The project was conceived by S.C., P.G., K.Z., W.W. and J.Z. M.G. and G.Y. synthesized HNC-1664 and 1664-TP. The in vitro antiviral effects of HNC-1664 were detected by X. Jia., E.L., Y.L., and M.Z. In vivo antiviral effects of HNC-1664 were detected by X. Jia., Y.L., and Y. Hou. Statistical analysis was performed by X. Jia., and Y.L. Enzymology assays of SARS-CoV-2 RdRP were performed by Q.L., M.L., L.A., S.L., and Y. Hong. Enzymology assays of LASV and EV71 RdRP were performed by X. Jing. and R.L., respectively. Cryo-EM experiments of SARS-CoV-2 and LASV RdRPs and crystallography experiments of EV71 RdRP were carried out by M.L., X. Jing., and Y.Z., respectively. The enzymology data analysis was performed by K.Z., M.L., S.L., R.L., Q.L., X. Jing., X. Jia., and P.G. The original manuscript was assembled by X. Jia., X. Jing., M.L., K.Z., P.G., and S.C., and all the authors edited and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Bruno Canard, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jia, X., Jing, X., Li, M. et al. An adenosine analog shows high antiviral potency against coronavirus and arenavirus mainly through an unusual base pairing mode. Nat Commun 15, 10750 (2024). https://doi.org/10.1038/s41467-024-54918-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54918-3