Abstract
Objective
To develop a population pharmacokinetic model for lopinavir boosted by ritonavir in coronavirus disease 2019 (Covid-19) patients.
Methods
Concentrations of lopinavir/ritonavir were assayed by an accredited LC-MS/MS method. The population pharmacokinetics of lopinavir was described using non-linear mixed-effects modeling (NONMEM version 7.4). After determination of the base model that better described the data set, the influence of covariates (age, body weight, height, body mass index (BMI), gender, creatinine, aspartate aminotransferase (AST), alanine aminotransferase (ALT), C reactive protein (CRP), and trough ritonavir concentrations) was tested on the model.
Results
From 13 hospitalized patients (4 females, 9 males, age = 64 ± 16 years), 70 lopinavir/ritonavir plasma concentrations were available for analysis. The data were best described by a one-compartment model with a first-order input (KA). Among the covariates tested on the PK parameters, only the ritonavir trough concentrations had a significant effect on CL/F and improved the fit. Model-based simulations with the final parameter estimates under a regimen lopinavir/ritonavir 400/100 mg b.i.d. showed a high variability with median concentration between 20 and 30 mg/L (Cmin/Cmax) and the 90% prediction intervals within the range 1–100 mg/L.
Conclusion
According to the estimated 50% effective concentration of lopinavir against SARS-CoV-2 virus in Vero E6 cells (16.7 mg/L), our model showed that at steady state, a dose of 400 mg b.i.d. led to 40% of patients below the minimum effective concentration while a dose of 1200 mg b.i.d. will reduce this proportion to 22%.
Similar content being viewed by others
Introduction
The pandemic of coronavirus disease 2019 (Covid-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) needs to identify effective drugs for treatment. Until now, no proven effective therapies for this virus has been highlighted. Remdesivir, a nucleoside analog, has shown effect on coronavirus in vitro [1, 2] but it is not still European Medicine Agency approved, even if it has been recently approved by US Food and Drug Administration. It is still currently being tested in ongoing randomized trials. So, it can be obtained only for a compassionate use at least in Europe [3]. Hydroxychloroquine has been proposed, in association or not with azithromycin, to treat Covid-19 patients, sometimes with success [4] and others with failure [5]. The association of lopinavir and ritonavir has been developed in order to treat human immunodeficiency virus (HIV), and it has been shown to have an excellent diffusion including in cerebrospinal fluid [6]. Lopinavir presented antiviral effect against SARS-CoV-2 virus in Vero E6 cells with a minimal effective concentration (MEC) at 26.63 μM (molecular weight = 628.8 g/mol, corresponding to 16.7 mg/L) in vitro [2]. Both drugs interacted well with the residues at the active site of the SARS-CoV-2 protease, and this interaction seemed to play an important role in the drug binding on the virus and the global effect of the drugs on the virus [7]. Although ritonavir is a protease inhibitor like lopinavir, it is used at low dose associated with lopinavir in order to inhibit cytochrome P450 3A4 (CYP3A4) isoenzyme which is responsible for the metabolism of lopinavir, leading to markedly increased plasma lopinavir concentrations [8, 9]. A ritonavir plasma concentration of 0.06 mg/L has been associated with a 50% maximum inhibition of the lopinavir clearance at the standard 400 mg lopinavir-100 mg ritonavir (LPV/r) twice-daily dose in healthy subjects [10] and 0.36 mg/L in HIV-infected patients [11]. A few clinical studies have been conducted in Covid-19 using the association LPV/r. Some precedent studies have shown positive results in severe acute respiratory syndrome with SARS-CoV-1 [12, 13]. However, a randomized, controlled, open-label trial was conducted in 199 adult patients hospitalized for severe and confirmed Covid-19 by Cao et al., showing no benefit with LPV/r treatment beyond standard care [14], leading many clinicians to abandon the use of this treatment. However, this study has been contested by many authors, for two main reasons: study statistically underpowered to demonstrate the chosen primary endpoint (time to clinical improvement) [15,16,17] and a median interval of 13 days between symptom onset and drug administration, which appeared to be too late since antiviral drugs were most effective when they were administered early in the infection [18,19]. In addition, in this study, no data on lopinavir exposure were available. These points of view led Cao et al. to reconsider their main conclusion and to declare that the association LPV/r could still be a potential therapeutic agent against Covid-19 [20]. However, one point could be critical and was not still studied: the variability in the pharmacokinetics (PK) of LPV/r in Covid-19 patients. Indeed, a study has shown that body weight was the only demographic factor that could be related to lopinavir exposure in HIV patient, leading clinicians to be aware for an increased risk of suboptimal antiviral efficacy in patients with high body weight [21]. Since a relationship seems to exist between critically ill Covid-19 patients and obesity [22,23], it will be important to determine the PK parameters of lopinavir in this population. So, the aim of this study was to evaluate the population PK of lopinavir in Covid-19 patients.
Methods
Patients
All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All patients were included in the RHU (Hospital-University Research in health) RECORDS program on sepsis (from ANR, French National Agency of Research) and gave their informed consent to participate to this program. All adults admitted to the intensive care unit (ICU) or in the medicine wards for a COVID-19 infection confirmed by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) reverse transcriptase-polymerase chain reaction (RT-PCR) and/or a compatible pulmonary computerized tomography-scan and treated with LPV/r (Kaletra®) with at least one sampling available (1 to 7/patient) for measurement of plasma concentrations of both compounds were retrospectively included in this study. All patients received 400 mg/100 mg of LPV/r twice daily.
LPV/r measurement
Sampling of blood for measurement of LPV/r was carried out at the start of treatment for 2 patients (no. 5 and 11). In all patients except one (no. 13, sampled 5 h after administration), the first sampling corresponded to a trough concentration, and if possible, sampling was carried out 2 h, 4 h, 6 h, 8 h, 10 h, and 12 h after drug administration. Blood was collected in sample tube containing EDTA as anticoagulant. After centrifugation and decantation, plasma samples were kept frozen at − 20 °C until analysis, which occurred in the next week. Plasma concentrations of lopinavir and ritonavir were quantified using a validated and accredited (COFRAC, Comité Français d’Accréditation) turbulent-flow liquid chromatography mass spectrometry method. Briefly, 50 μL of sample, calibrator, or quality control was treated with 75 μL of methanol (precipitation reagent) containing internal standard (lopinavir-D8). Samples were immediately vortexed, stored 15 min at 4 °C, and centrifuged at 14,000g for 15 min. The supernatant was injected into the chromatographic system, consisting on a Turboflow® on-line extraction system using a cyclone column (50 × 0.5 mm, 50 μm particles), a CTC auto sampler, and a triple quadruple mass spectrometer TSQ QUANTUM Access MAX equipped with an electrospray ionization interface. The analytical HPLC column was a Hypersil GOLD (50 × 3 mm × 3 μm) column. Data analysis was performed using an LCQuan™ 2.7 software package (all Thermo Fisher Scientific®, Les Ulis, France).
Population pharmacokinetic analysis
The population PK analysis of lopinavir plasma concentrations was performed using the software non-linear mixed-effects modeling version 7.4 (NONMEM Icon Development Solutions, Ellicott City, MD). The usual first-order conditional estimation (FOCE) method with the interaction option was used throughout. Data management, statistical analysis, and graphical outputs were realized with R version 3.6.1. [24].
For the structural model, the NONMEM analysis subroutines ADVAN2 and ADVAN 4 were used to test for one- and two-compartment models with an oral dosing compartment. To describe the oral absorption phase, zero-order and first-order absorption models were tested in addition to a lag-time. Inter-subject variability of the different PK parameters was estimated with a proportional error model. Several error models (additive, proportional, or both) were investigated to describe residual variability. The performance of the model was judged by both statistical and graphic methods [25,26]. The minimal value of the objective function (OFV) as calculated by NONMEM was also used to assess the goodness-of-fit. An increase in goodness-of-fit is accompanied by a decrease in objective function, and this decrease is asymptotically distributed as a chi-square distribution. Furthermore, relative standard errors (RSE) were calculated by use of the COVARIANCE option of NONMEM. For graphic model diagnostics, the following graphs were compared (“goodness-of-fit plots” or “diagnostic plots”): observed concentrations (depending variable, DV) vs. predictions and normalized predictive distribution error (NPDE) vs. predictions or TIME [25,26].
This first analysis was performed to define the base model that better described the data set. Once this has been realized, the influence of each covariate was tested on the model. These covariates were age, body weight, height, body mass index (BMI), gender, creatinine, aspartate aminotransferase (AST), alanine aminotransferase (ALT), C reactive protein (CRP), and trough ritonavir concentrations. The potential influence of the covariates was tested using linear or non-linear function as appropriate. The continuous variables were centered on their median value and categorical variables were coded as 0 or 1. The effect of ritonavir trough concentrations on lopinavir clearance was further tested with the following equation:
where CL/F is the lopinavir clearance, CL0/F the lopinavir clearance in the absence of ritonavir, Imax the maximum inhibitory effect of ritonavir on CL/F; CresRTV the trough concentration of ritonavir (mg/mL), and IC50 the ritonavir concentration producing half of the Imax.
The diagnostic plots described above, the change in objective function, and the change in parameter variability were noted to select the covariates which improved the model prediction. A decrease in the objective function value (OFV) of at least 3.84 (chi-squared distribution with one degree of freedom for P < 0.05) relative to the base PK model was required for the addition of a single parameter in the model. All selected covariates were included in a so-called full model. Then a backward elimination procedure was performed in which each covariate was removed in turn from the full model and the difference in OFV between the full and each reduced model was examined. An increase in OFV greater than 6.63 (P < 0.01, 1 degree of freedom) was required to retain the covariate in the final model.
The graphical evaluation described above as diagnostic plots were performed for all models as well as for the final model. The precision of the parameter estimates was expressed as relative standard error (RSE, %) and confidence intervals (CI). The RSE were directly computed by NONMEM, and a value < 30% for fixed effects and < 50% for random effects were considered acceptable. A bootstrap method was used to verify the robustness of standard approximations for parameter uncertainty. The percentile bootstrap confidence intervals were constructed by taking the lower 2.5% and the upper 97.5% value of each parameter. To perform a prediction-corrected visual predictive check (pcVPC), the final model, the corresponding parameter values (including inter-individual variability and residual variability), and the original dataset were used to simulate 500 replicates, using the SIMULATION, SUBPROBLEM feature in NONMEM (Monte Carlo simulation).
Simulations of 500 individuals based on the final model with between subject variability were conducted to describe the expected time course of lopinavir concentrations during the whole treatment. Another set of simulations was performed to explore new dosing options. Using the final model with the corresponding estimated parameters, Monte Carlo simulation (n = 500) predicted the doses reaching the MEC at steady state and the proportion of patients below this threshold. An empirical cumulative distribution function was computed using the R software [24].
Results
Thirteen patients were included: 4 females and 9 males, age = 64 ± 16 years, body weight = 85 ± 15 kg, and body mass index (BMI) = 27.9 ± 5.4 kg/m2. Demographic and clinical characteristics of these patients are detailed in Table 1.
The data of plasma lopinavir concentrations obtained in the thirteen patients according to the day of treatment and their evolution in terms of survival are shown in Fig. 1. The trough ritonavir concentrations were in the range < 0.02–1.5 mg/L. Different structural PK models were tested, one compartment and two compartments, and the lopinavir concentrations were best described by a one-compartment model with a first-order input (KA), a central volume of distribution (V/F), and a clearance (CL/F). However, the first-order absorption rate constant (KA) was not estimable appropriately, and the relative standard error (RSE) was higher than 50%. It was then decided to fix the KA to a constant value. Several values of KA were tested to evaluate the model sensitivity. A value close to a previous published KA value obtained in healthy volunteers [10] allowed the better estimation of other parameters and was retained for the analysis. The addition of a lag time failed to produce standard error estimates and was not included. The inter-individual variability (IIV) was estimated on clearance (CL/F) and on the volume of distribution (V/F). The residual unexplained variability (RUV) was modeled as proportional plus a fixed additive part. Among the covariates tested on the PK parameters, only the ritonavir trough concentrations had a significant effect on CL/F and improved the fit. The best model was a direct relationship between ritonavir trough concentrations and CL/F with a maximal inhibitory effect (Eq. 1). Owing to identifiability problems (RSE > 50%), the theta values for Imax and IC50 were fixed. After testing for different values, the more precise estimation of other parameters was found by using previous published values of Imax and IC50 [10]. The final population PK estimates of other parameters were well described with a small relative standard errors (RSE < 30%) and within the 95% confidence intervals constructed by the bootstrap. These results are presented in Table 2 together with the shrinkage estimates for CL/F and V/F. The diagnostic plots (supplemental Fig I) as well as the pcVPC (Fig. 2) did not show a trend, indicating that the model adequately describes the PK profile of lopinavir. Model-based simulations were performed with the final parameter estimates under a similar regimen, lopinavir 400 mg every 12 h (plus ritonavir b.i.d.). The median concentrations and the 90% prediction interval are presented with the MEC of lopinavir against SARS-CoV-2 virus in Vero E6 cells (Fig. 3). An empirical cumulative distribution function (ECDF) depicted the percent of patients below the MEC according to doses ranging from 400 to 1200 mg every 12 h (Fig. 4). The figure showed that a dose of 400 mg b.i.d. at steady state led to 40% of patients below the MEC while a dose of 1200 mg b.i.d. reduced this proportion to 22%.
Prediction-corrected visual predictive check of the final model of lopinavir. Open circles represent prediction-corrected observed concentrations; black solid line and dashed lines represent the median and the 90% predicted interval (90%PI) of the observed data; shaded area represent the model-predicted 95% confidence interval of the simulated median and 90%PI
Concentration versus time of lopinavir simulations over 10 days following 400 mg bid in covid-19 patients. Under the figure: The continuous line represents the median lopinavir concentration and the shaded area the 90% prediction interval determined by 500 simulations (5th/95th). The horizontal dashed line is the estimated minimal effective concentration (MEC) of lopinavir (16.7 mg/L) against SARS-CoV-2 virus in Vero E6 cells [2]
Discussion
In this study, we were able to model the population PK of lopinavir in 13 Covid-19 treated patients using 70 measurements of plasma concentration of lopinavir in these patients. Two main points appeared in the study: the high variability of lopinavir concentrations observed in the Covid-19 patients and the median concentrations obtained at steady state which appeared to be near the estimated 50% effective values of lopinavir concentration against SARS-CoV-2 virus in vitro in Vero E6 cells [2].
The limited sample size (13 patients only) and the sparse sampling in some of them did not allow to estimate all the PK parameters, which was a limitation of our study. However, we decided to fix the KA and the theta values for Imax and IC50 according to previous published PK values in healthy subjects [10], and we controlled to allow a more precise estimation of other parameters by testing different values.
Despite this limitation, a population PK model for lopinavir/ritonavir used in Covid-19 patients was developed giving an adequate description of the PK of lopinavir. A relationship between the apparent clearance CL/F of lopinavir and the exposure to ritonavir was found, as previously shown in healthy volunteers [10] and in human immunodeficiency virus (HIV) type-1 infected patients [27], in relation with the inhibitory effect of ritonavir on CYP3A4 activity. It has been shown that this inhibitory effect on the metabolism of lopinavir by ritonavir was obtained with very low concentration of ritonavir, with IC50 values of 0.05 mg/L on human liver microsomes [28] and 50% of lopinavir CL/F inhibition with ritonavir plasma concentration of 0.06 mg/L in healthy volunteers [10] or 0.36 mg/L in HIV-1 patients [11]. Only 13 samples among the 70 samples collected in the study had concentrations below this cut-off, where 7 of those samples were from the same patient (no. 11) with all values < 0.02 mg/L (with lopinavir concentrations also very low, all < 0.07 mg/L). CL/F of lopinavir without ritonavir in Covid-19 patients (4.88 L/h) appeared to be lower than those obtained in healthy volunteers which was found at 21.6 L/h [10] and 14.8 L/h in HIV-patients [9]. This lower CL/F is probably due to the characteristics of our patients since 10 of our 13 patients were in ICU, intubated, and ventilated. Lopinavir (and ritonavir) are metabolized by CYP3A4. Infection and inflammation are associated with downregulation of these cytochrome [29], and lopinavir concentrations are modified by the blood CRP levels [30]. Our patients were infected by severe acute respiratory syndrome coronavirus 2, and all of them presented high level of inflammation. At the moment of sampling for lopinavir/ritonavir plasma concentrations, all had CRP levels higher than 100 mg/L except patient no. 3 and 7 where CRP was available only 2 days after sampling (with level at 34 mg/L and 90 mg/L, normal value < 5 mg/L), which could then explain the decrease in apparent clearance observed in our patients. Moreover, ten patients among the 13 had renal failure, with creatinine > 104 μmol/L and until 478 μmol/L (normal range of the laboratory = 59–104 μmol/L) which could decrease the renal elimination of the compounds, even if the excretion of unchanged lopinavir in urine is low. V/F was found at 94.8 L in our patients, which is higher than the values observed in healthy subjects (55.3 L) [10] or in HIV-infected patients (61.6 L) [27]. This difference could be also due to the characteristics of our patients since 8 were in overweight (BMI > 25), and 4 were obese (BMI > 30). The effect of weight on lopinavir concentrations has already been reported by Stohr et al. [31]. The absorption rate constant was fixed in our study at 0.572 h−1, since values found in other studies in HIV-1 infected patients (0.564 h−1) [27] or healthy subjects (0.57 h−1) [10] were similar. However, 10 of our patients were in ICU and tablets were administered by nasogastric tube after crushing them, which could have modified this absorption rate.
The concentrations measured in the patients showed very large variation, with 45 mg/L in one patient (patient no. 9) after 1 day of treatment to 0.7 mg/L (no. 7) after 5 days or 0.9 mg/L (no. 4) after 3 days. This variation could be due in part to a poor intestinal absorption of lopinavir in some patients (patient no. 11 for example) and to the decrease clearance in some others. The high variation of concentrations did not seem to be correlated with survival (Fig. 1), since 2 patients with very high lopinavir concentrations died 6 and 11 days after sampling (patient no. 8 and 9, respectively), and 2 patients with very low concentrations stayed alive and have recovered from their illness (patient no. 4 and 7). The median plasma lopinavir concentrations predicted with our model at steady state was between 20 and 30 mg/L, with large 90% prediction interval between 1 and 100 mg/L (Fig. 3). Median (range) trough concentrations in the same dosage scheme than our patients were found in Covid-19 patients at 18 mg/L (11.4 to 30.8 mg/L) by other authors, showing also a high variation of concentrations [32]. This median plasma lopinavir concentration observed in Covid-19 patients is high compared to that usually observed in HIV-1 infected patients, around 5 to 6 mg/L at the same dosage regimen (400/100 mg b.i.d) [33], probably related to the decrease apparent clearance highlighted in our model. However, the median concentrations obtained appeared to be near the estimated MEC of lopinavir against SARS-CoV-2 virus in vitro in Vero E6 cells (16.7 mg/L) [2]. That means that near 50% of the patients did not achieve enough plasma concentration in order to fight the virus. Moreover, the concentration of lopinavir has been shown to be around 1.7 higher in epithelial lining fluid than in plasma in one patient, which means that probably concentrations obtained in lung in our patient should be at least for half of the patients under the active concentration on SARS-CoV-2 virus [34]. Using our model, we evaluated that a dose of 1200 mg b.i.d. will reduce this proportion to 22%. A probably good strategy could be to have a 400/100 mg as a starting dose, to use therapeutic drug monitoring after collecting a trough plasmatic concentration rapidly (around 3 days after first administration according Fig. 3) and, if below the targeted effective concentration, to increase until 1200/400 mg b.i.d. which will decrease the number of patient below the effective concentration. However, side effects at such a dose remain to be assessed.
In conclusion, our model-based simulations in Covid-19 patients with the final parameter estimates under a regimen lopinavir/ritonavir 400/100 mg b.i.d. showed a high variability with median concentration between 20 and 30 mg/L (Cmin/Cmax) and the 90% prediction interval = 1–100 mg/L. According to the estimated MEC of lopinavir against SARS-CoV-2 virus in Vero E6 cells (16.7 mg/L), near 50% of patients did not reach enough concentration in plasma and lung with a classical regimen lopinavir/ritonavir 400/100 mg b.i.d.
References
Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, Smith EC, Case JB, Feng JY, Jordan R, Ray AS, Cihlar T, Siegel D, Mackman RL, Clarke MO, Baric RS, Denison MR (2018) Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 9(2):e00221–e00218. https://doi.org/10.1128/mBio.00221-18
Choy KT, Yin-Lam Wong A, Kaewpreedee P, Sia SF, Chen D, Yan Hui KP, Wing Chu DK, Wai Chan MC, Pak-Hang Cheung P, Huang X, Peiris M, Yen HL (2020) Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir Res 104786:104786. https://doi.org/10.1016/j.antiviral.2020.104786
Lapadula G (2020) Five reasons why data on compassionate use of remdesivir deserved publication (and are worth reading). Swiss Med Wkly 150:w20265. https://doi.org/10.4414/smw.2020.20265
Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira VE, Dupont HT, Honoré S, Colson P, Chabrière E, La Scola B, Rolain JM, Brouqui P, Raoult D (2020) Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 105949:105949. https://doi.org/10.1016/j.ijantimicag.2020.105949
Mehra MR, Desai SS, Ruschitzka F, Patel AN (2020) Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis. In: Lancet. https://doi.org/10.1016/S0140-6736(20)31180-6
de Truchis P, Mathez D, Abe E, Dinh A, Ledu D, Greder-Belan A, Alvarez JC (2010) Cerebrospinal fluid HIV-1 virological escape with lymphocytic meningitis under lopinavir/ritonavir monotherapy. AIDS. 24(8):1235–1236
Nutho B, Mahalapbutr P, Hengphasatporn K, Pattaranggoon NC, Simanon N, Shigeta Y, Hannongbua S, Rungrotmongkol T (2020) Why are lopinavir and ritonavir effective against the newly emerged coronavirus 2019? Atom Insig Inhibit Mechan Biochem 59(18):1769–1779
Zeldin RK, Petruschke RA (2004) Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J Antimicrob Chemother 53(1):4–9
Crommentuyn KM, Kappelhoff BS, Mulder JW, Mairuhu AT, van Gorp EC, Meenhorst PL, Huitema AD, Beijnen JH (2005) Population pharmacokinetics of lopinavir in combination with ritonavir in HIV-1-infected patients. Br J Clin Pharmacol 60(4):378–389
Dickinson L, Boffito M, Back D, Else L, von Hentig N, Davies G, Khoo S, Pozniak A, Moyle G, Aarons L (2011) Sequential population pharmacokinetic modeling of lopinavir and ritonavir in healthy volunteers and assessment of different dosing strategies. Antimicrob Agents Chemother 55(6):2775–2782
Moltó J, Barbanoj MJ, Miranda C, Blanco A, Santos JR, Negredo E, Costa J, Domingo P, Clotet B, Valle M (2008) Simultaneous population pharmacokinetic model for lopinavir and ritonavir in HIV-infected adults. Clin Pharmacokinet 47(10):681–692
Chu CM, Cheng VC, Hung IF, Wong MM, Chan KH, Chan KS, Kao RY, Poon LL, Wong CL, Guan Y, Peiris JS, Yuen KY, HKU/UCH SARS study group (2004) Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 59(3):252–256
Chan KS, Lai ST, Chu CM, Tsui E, Tam CY, Wong MM, Tse MW, Que TL, Peiris JS, Sung J, Wong VC, Yuen KY (2003) Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicentre retrospective matched cohort study. Hong Kong Med J 9(6):399–406
Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, Li X, Xia J, Chen N, Xiang J, Yu T, Bai T, Xie X, Zhang L, Li C, Yuan Y, Chen H, Li H, Huang H, Tu S, Gong F, Liu Y, Wei Y, Dong C, Zhou F, Gu X, Xu J, Liu Z, Zhang Y, Li H, Shang L, Wang K, Li K, Zhou X, Dong X, Qu Z, Lu S, Hu X, Ruan S, Luo S, Wu J, Peng L, Cheng F, Pan L, Zou J, Jia C, Wang J, Liu X, Wang S, Wu X, Ge Q, He J, Zhan H, Qiu F, Guo L, Huang C, Jaki T, Hayden FG, Horby PW, Zhang D, Wang C (2020) A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med 382(19):1787–1799
Dalerba P, Levin B, Thompson JL (2020) A Trial of Lopinavir-Ritonavir in Covid-19. N Engl J Med 382(21):e68. https://doi.org/10.1056/NEJMc2008043
Corrao S, Natoli G, Cacopardo B (2020) A Trial of Lopinavir-Ritonavir in Covid-19. N Engl J Med 382(21):e68. https://doi.org/10.1056/NEJMc2008043
Carmona-Bayonas A, Jimenez-Fonseca P, Castañón E (2020) A Trial of Lopinavir-Ritonavir in Covid-19. N Engl J Med 382(21):e68. https://doi.org/10.1056/NEJMc2008043
Kunz KM (2020) A Trial of Lopinavir-Ritonavir in Covid-19. N Engl J Med 382(21):e68. https://doi.org/10.1056/NEJMc2008043
Havlichek D Jr (2020) A Trial of Lopinavir-Ritonavir in Covid-19. N Engl J Med 382(21):e68. https://doi.org/10.1056/NEJMc2008043
Cao B, Zhang D, Wang C (2020) A Trial of Lopinavir-Ritonavir in Covid-19. N Engl J Med 382(21):e68. https://doi.org/10.1056/NEJMc2008043
van der Leur MR, Burger DM, la Porte CJ, Koopmans PP (2006) A retrospective TDM database analysis of interpatient variability in the pharmacokinetics of lopinavir in HIV-infected adults. Ther Drug Monit 28(5):650–653
Buscemi S, Buscemi C, Batsis JA. There is a relationship between obesity and COVID-19 but more information is needed. Obesity (Silver Spring). 2020 May 12. doi: https://doi.org/10.1002/oby.22883
Petrakis D, Margină D, Tsarouhas K, Tekos F, Stan M, Nikitovic D, Kouretas D, Spandidos DA, Tsatsakis A (2020) Obesity - a risk factor for increased COVID-19 prevalence, severity and lethality (review). Mol Med Rep 22:9–19. https://doi.org/10.3892/mmr.2020.11127
Core Team R (2019) R: a language and environment for statistical computing. In: R Foundation for statistical computing. Austria, Vienna https://www.R-project.org/
Mould DR, Upton R (2013) Basic concepts in population modeling, simulation, and model-based drug development—part 2: introduction to pharmacokinetic modeling methods. CPT Pharmacometrics Syst Pharmacol 2:e38. https://doi.org/10.1038/psp.2013.14
Brendel K, Comets E, Laffont C, Laveille C, Mentre F (2006) Metrics for external model evaluation with an application to the population pharmacokinetics of gliclazide. Pharm Res 23:2036–2049
Crommentuyn KM, Mulder JW, Mairuhu AT, van Gorp EC, Meenhorst PL, Huitema AD, Beijnen JH (2004) The plasma and intracellular steady-state pharmacokinetics of lopinavir/ritonavir in HIV-1-infected patients. Antivir Ther 9(5):779–785
Sham HL, Kempf DJ, Molla A, Marsh KC, Kumar GN, Chen CM, Kati W, Stewart K, Lal R, Hsu A, Betebenner D, Korneyeva M, Vasavanonda S, McDonald E, Saldivar A, Wideburg N, Chen X, Niu P, Park C, Jayanti V, Grabowski B, Granneman GR, Sun E, Japour AJ, Leonard JM, Plattner JJ, Norbeck DW (1998) ABT-378, a highly potent inhibitor of the human immunodeficiency virus protease. Antimicrob Agents Chemother 42(12):3218–3224
Morgan ET (2009) Impact of infectious and inflammatory disease on cytochrome P450-mediated drug metabolism and pharmacokinetics. Clin Pharmacol Ther 85(4):434–438
Ofotokun I, Lennox JL, Eaton ME, Ritchie JC, Easley KA, Masalovich SE, Long MC, Acosta EP (2011) Immune activation mediated change in alpha-1-acid glycoprotein: impact on total and free lopinavir plasma exposure. J Clin Pharmacol 51(11):1539–1548
Stöhr W, Back D, Dunn D, Sabin C, Winston A, Gilson R, Pillay D, Hill T, Ainsworth J, Gazzard B, Leen C, Bansi L, Fisher M, Orkin C, Anderson J, Johnson M, Easterbrook P, Gibbons S, Khoo S, UK CHIC Steering Committee (2010) Factors influencing lopinavir and atazanavir plasma concentration. J Antimicrob Chemother 65(1):129–137
Gregoire M, Le Turnier P, Gaborit BJ, Veyrac G, Lecomte R, Boutoille D, Canet E, Imbert BM, Bellouard R, Raffi F (2020) Lopinavir pharmacokinetics in COVID-19 patients. J Antimicrob Chemother 75:2702–2704. https://doi.org/10.1093/jac/dkaa195
Boffito M, Back DJ, Hoggard PG, Caci A, Bonora S, Raiteri R, Sinicco A, Reynolds HE, Khoo S, Di Perri G (2003) Intra-individual variability in lopinavir plasma trough concentrations supports therapeutic drug monitoring. AIDS. 17(7):1107–1108
Atzori C, Villani P, Regazzi M, Maruzzi M, Cargnel A (2003) Detection of intrapulmonary concentration of lopinavir in an HIV-infected patient. AIDS. 17(11):1710–1711
Acknowledgements
We thank The Garches COVID 19 Collaborative Group. Department of intensive care: AMBROSI, Xavier, MD; AMTHOR Suzanne, MD; BOUNAB, Rania, MD; CHENTOUH, Ryme, MD; CLAIR, Bernard, MD; FAYSSOIL, Abdallah, MD; FRIEDMAN, Diane, MD; HEMING, Nicholas, MD; MAXIME, Virginie, MD, NIEL DURIEZ, Myriam, MD; ORLIKOWSKI, David, MD, PhD; SANTI, Francesca, MD. Pharmacy: VILLART, Maryvonne, PharmD, MICHELON Hugues, PharmD, MSc. Resident of the Department of Intensive Care: ABBAR, Baptiste, Interne; DRAY, Juliah, Interne; TAMAYO, Juan, Interne; PASCAULT, Alice, Interne; ZINI; Justine, Interne; BENNINGTON, Steven, Interne; MOUCACHEN, Myriam, Interne; GAY, Pierre, Interne; LUXMAN, Majistor, Interne; KOCHBATI, Elias, Interne. Department of Anesthesia, Perioperative Care, and Pain: MARTINEZ, Valéria, MD, Ph D; GUICHARD Léa, MD; TRABELSI, Chawki, MD; BOUTROS, Marie, MD; SCHITTER Sebastien, MD; MEULEYE, Simone, MD; REYSZ, Suzanne, MD; KHITER, Hakim, MD; DOSNE BLACHIER, Brigitte, MD; BRON, Anne Lyse, MD; DEFOUCHECOUR Etiennette, MD; HAMON PIETRIN, Damien, MD, PhD;
COESTER, Denys, MD. Pediatric Critical Care Unit: BERGOUNIOUX Jean MD; OMAR Amal, MD; GUILLON Maud, MD; AMTHOR Helge, MD, PhD. Laboratory of Physiology: PRIGENT Helene, MD PhD; LOFASO Frédéric, MD PhD. Department of Rehabilitation and Physical Medicine: DENYS Pierre, MD, PhD; BENSMAIL, Djamel, MD PhD; JOUSSAIN Charles, MD PhD; MALOT Claire, MD; LANSAMAN Thibaut, MD; LEOTARD Antoine, MD MS; LE LIEPVRE Hélène, MD; RECH Celia, MD; PAQUEREAU Julie, MD; KAGANE Lauren, MD; LEVY Jonathan MD MS. Resident–Department of Rehabilitation: CHKRON Elsa, interne; ANGIONI Florence, interne; KARABULUT Céline, interne; LEMOINE Jérôme, interne; VIBERT Julien interne; TRYSTRAM Noémie, interne. Department of Infectious Diseases: Christian PERRONNE, MD, PhD; Véronique PERRONNE, MD; Aurélien DINH, MD, MS; Pierre de TRUCHIS, MD, MS; Soline SIMEON, MD, MS; Simon BESSIS, MD, Morgan MATT, MD; Hélène MASCITTI, MD, MS; Stéphanie LANDOWSKI, MD;
Pascal CRENN, MD, PhD. Resident of Department of Infectious Diseases: Aurélie LE GAL, interne; Aymeric LANORE, interne; Julia NGUYEN VAN THANG, interne; Louis JACOB, interne; Nicolas KIAVUE, interne; Marc HOBEIKA, interne. Department of Microbiology and Virology: Jean Louis GAILLARD, MD, PhD; Jean Louis HERMANN, MD, PhD; Martin ROTTMAN, MD, PhD; Anne Laure ROUX, MD, PhD; Marie-Anne WELTI, MD, PhD; Elyanne GAULT, MD, PhD; Christine LAWRENCE.
Author information
Authors and Affiliations
Consortia
Contributions
JCA wrote the paper and managed all data; PM, DA and BD included and investigated the patients; NS managed data; IET and IAL managed samples and analysis for lopinavir/ritonavir.
Corresponding author
Ethics declarations
All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All patients were included in the RHU (Hospital-University Research in health) RECORDS program on sepsis (from ANR, French National Agency of Research) and gave their informed consent to participate to this program.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
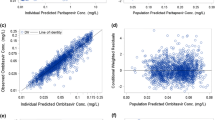
Fig. I
Goodness-of-plots for the final lopinavir population pharmacokinetic model. 2a: Observed concentrations (DV) versus population predictions (PRED), 2b: Observed concentrations versus individual predictions (IPRED), 2c: normalized predictive distribution error (NPDE) versus predictions, 2d: NPDE vs TIME. Under the figure: The line of identity (black line) and the linear regression (dotted line) are shown. (PPTX 68 kb)
Rights and permissions
About this article
Cite this article
Alvarez, J.C., Moine, P., Davido, B. et al. Population pharmacokinetics of lopinavir/ritonavir in Covid-19 patients. Eur J Clin Pharmacol 77, 389–397 (2021). https://doi.org/10.1007/s00228-020-03020-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00228-020-03020-w