Abstract
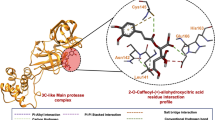
Since the coronavirus disease 2019 (COVID-19) outbreak, research has been conducted on treatment and countermeasures against the causative severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, the development of new seeds is urgently needed because viruses have the characteristic of becoming resistant through mutation. We hypothesize that endophytes produce antiviral substances to combat foreign viruses in host plants. According to this hypothesis, the seeds of therapeutic agents for infectious diseases could be obtained from endophytes by culture experiments. This report found that Aspergillus sp. endophyte isolated from Catharanthus roseus produced ( +)-eupenoxide and its 3-ketone form with anti-SARS-CoV-2 activity. In addition, ( +)-eupenoxide-3,6-diketon was discovered as a new compound with potent 3C-like protease inhibitory activity (IC50: 0.048 μM) by synthesis based on ( +)-eupenoxide. This finding could be an important evidence that endophytic fungi symbiosis with medicinal plants is useful as antiviral producers.
Graphical abstract










Similar content being viewed by others
References
Wani ZA, Ashraf N, Mohiuddin T, Riyaz-Ul-Hassan S (2015) Plant-endophyte symbiosis, an ecological perspective. Appl Microbiol Biotechnol 99:2955–2965. https://doi.org/10.1007/S00253-015-6487-3/FIGURES/3
Saikkonen K, Wäli P, Helander M, Faeth SH (2004) Evolution of endophyte–plant symbioses. Trends Plant Sci 9:275–280. https://doi.org/10.1016/J.TPLANTS.2004.04.005
Arachevaleta M, Bacon CW, Hoveland CS, Radcliffe DE (1989) Effect of the tall fescue endophyte on plant response to environmental stress. Agron J 81:83–90. https://doi.org/10.2134/agronj1989.00021962008100010015x
Nejidat A (1987) Effect of ophiobolin A on stomatal movement: role of calmodulin. Plant Cell Physiol 28:455–460
Kanda K, Hirai Y, Koga H, Hasegawa K (1994) Endophyte-enhanced resistance in perennial ryegrass and tall fescue to bluegrass webworm, parapediasia teterrella. Japanese J Appl Entomol Zool 38:141–145. https://doi.org/10.1303/jjaez.38.141
Trivedi P, Leach JE, Tringe SG et al (2020) Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 1811(18):607–621. https://doi.org/10.1038/s41579-020-0412-1
Maehara S, Nakajima S, Watashi K et al (2023) Anti-SARS-CoV-2 agents in artemisia endophytic fungi and their abundance in artemisia vulgaris tissue. J Fungi 9:905. https://doi.org/10.3390/jof9090905
Kumar A, Patil D, Rajamohanan PR, Ahmad A (2013) Isolation, purification and characterization of vinblastine and vincristine from endophytic fungus Fusarium oxysporum isolated from Catharanthus roseus. PLoS One. https://doi.org/10.1371/JOURNAL.PONE.0071805
Duke RK, Rickards RW (1984) Stereospecific total synthesis of the cyclohexene oxide antibiotic eupenoxide. J Org Chem 49:1898–1904. https://doi.org/10.1021/JO00185A010/ASSET/JO00185A010.FP.PNG_V03
Mehta G, Roy S, Davis RA (2008) On the stereostructures of (+)-eupenoxide and (−)-3′,4′-dihydrophomoxide: a caveat on the spectral comparisons of oxygenated cyclohexenoids. Tetrahedron Lett 49:5162–5164. https://doi.org/10.1016/J.TETLET.2008.06.076
Myobatake Y, Takemoto K, Kamisuki S et al (2014) Cytotoxic alkylated hydroquinone, phenol, and cyclohexenone derivatives from aspergillus violaceofuscus Gasperini. J Nat Prod 77:1236–1240. https://doi.org/10.1021/NP401017G/SUPPL_FILE/NP401017G_SI_001.PDF
Li C, Porco JA (2005) Synthesis of epoxyquinol A and related molecules: probing chemical reactivity of epoxyquinol dimers and 2H-pyran precursors. J Org Chem 70:6053–6065. https://doi.org/10.1021/JO050897O
Mehta G, Roy S (2004) Enantioselective total synthesis of (+)-eupenoxide and (+)-phomoxide: revision of structures and assignment of absolute configuration. Org Lett 6:2389–2392. https://doi.org/10.1021/OL0492288/SUPPL_FILE/OL0492288SI20040604_101136.PDF
Kleinke AS, Li C, Rabasso N, Porco JA (2006) Total synthesis of the interleukin-1β converting enzyme inhibitor EI-1941-2 using tandem oxa-electrocyclization/oxidation. Org Lett 8:2847–2850. https://doi.org/10.1021/ol060954f
Kakeya H, Miyake Y, Shoji M et al (2003) Novel non-peptide inhibitors targeting death receptor-Mediated apoptosis. Bioorg Med Chem Lett 13:3743–3746. https://doi.org/10.1016/J.BMCL.2003.08.003
Fujita K, Ishikawa F, Kakeya H (2014) Biosynthetic origins of the epoxyquinone skeleton in epoxyquinols A and B. J Nat Prod 77:2707–2710. https://doi.org/10.1021/NP5004615/SUPPL_FILE/NP5004615_SI_001.PDF
Li P, Takei R, Takahashi K, Nabeta K (2007) Biosynthesis of theobroxide and its related compounds, metabolites of Lasiodiplodia theobromae. Phytochemistry 68:819–823. https://doi.org/10.1016/J.PHYTOCHEM.2006.12.006
Tsukamoto S, Hirota H, Imachi M et al (2005) Himeic acid A: a new ubiquitin-activating enzyme inhibitor isolated from a marine-derived fungus, Aspergillus sp. Bioorg Med Chem Lett 15:191–194. https://doi.org/10.1016/J.BMCL.2004.10.012
Sakekar AA, Gaikwad SR, Punekar NS (2021) Protein expression and secretion by filamentous fungi. J Biosci 461(46):1–18. https://doi.org/10.1007/S12038-020-00120-8
Uzaki M, Mori T, Sato M et al (2024) Integration of cell differentiation and initiation of monoterpenoid indole alkaloid metabolism in seed germination of Catharanthus roseus. New Phytol 242:1156–1171. https://doi.org/10.1111/NPH.19662
Davis RA, Andjic V, Kotiw M, Shivas RG (2005) Phomoxins B and C: polyketides from an endophytic fungus of the genus Eupenicillium. Phytochemistry 66:2771–2775. https://doi.org/10.1016/J.PHYTOCHEM.2005.09.004
Dramae A, Bunbamrung N, Intaraudom C et al (2024) Polyoxygenated cyclohexenoids from the endophytic fungus, Aspergillus aculeatus BCC69431 and biological properties. Tetrahedron 150:133753. https://doi.org/10.1016/J.TET.2023.133753
Ohashi H, Watashi K, Saso W et al (2021) Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience. https://doi.org/10.1016/j.isci.2021.102367
Liu H, Iketani S, Zask A et al (2022) Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19. Nat Commun 131(13):1–16. https://doi.org/10.1038/s41467-022-29413-2
Ramos-Guzmán CA, Ruiz-Pernía JJ, Tuñón I (2021) Inhibition mechanism of SARS-CoV-2 main protease with ketone-based inhibitors unveiled by multiscale simulations: insights for improved designs**. Angew Chemie Int Ed 60:25933–25941. https://doi.org/10.1002/ANIE.202110027
Unoh Y, Uehara S, Nakahara K et al (2022) Discovery of S-217622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J Med Chem 65:6499–6512. https://doi.org/10.1021/ACS.JMEDCHEM.2C00117
Bai B, Arutyunova E, Khan MB et al (2021) Peptidomimetic nitrile warheads as SARS-CoV-2 3CL protease inhibitors. RSC Med Chem 12:1722–1730. https://doi.org/10.1039/D1MD00247C
Gao S, Sylvester K, Song L et al (2022) Discovery and crystallographic studies of trisubstituted piperazine derivatives as non-covalent SARS-CoV-2 main protease inhibitors with high target specificity and low toxicity. J Med Chem 65:13343–13364. https://doi.org/10.1021/ACS.JMEDCHEM.2C01146/SUPPL_FILE/JM2C01146_SI_002.CSV
Liu H, Ye F, Sun Q et al (2020) Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. bioRxiv. https://doi.org/10.1101/2020.04.10.035824
Shi TH, Huang YL, Chen CC et al (2020) Andrographolide and its fluorescent derivative inhibit the main proteases of 2019-nCoV and SARS-CoV through covalent linkage. Biochem Biophys Res Commun 533:467–473. https://doi.org/10.1016/J.BBRC.2020.08.086
Maehara S, Fathoni A, Tagawa M et al (2023) Environmental differences between Japan and Indonesia provide endophyte diversity associated with Artemisia plant and variety of artemisinin derivatives in microbial conversion. J Nat Med 77:916–927. https://doi.org/10.1007/s11418-023-01709-7
Pracht P, Grimme S, Bannwarth C et al (2024) CREST-a program for the exploration of low-energy molecular chemical space. J Chem Phys. https://doi.org/10.1063/5.0197592
Bannwarth C, Ehlert S, Grimme S (2019) GFN2-xTB-an accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions. J Chem Theory Comput 15:1652–1671. https://doi.org/10.1021/ACS.JCTC.8B01176
Grimme S, Bohle F, Hansen A et al (2021) Efficient quantum chemical calculation of structure ensembles and free energies for nonrigid molecules. J Phys Chem A 125:4039–4054. https://doi.org/10.1021/ACS.JPCA.1C00971
Neese F (2012) The ORCA program system. Wiley Interdiscip Rev Comput Mol Sci 2:73–78. https://doi.org/10.1002/WCMS.81
Neese F (2018) Software update: the ORCA program system, version 4.0. Wiley Interdiscip Rev Comput Mol Sci 8:e1327. https://doi.org/10.1002/WCMS.1327
Neese F, Wennmohs F, Becker U, Riplinger C (2020) The ORCA quantum chemistry program package. J Chem Phys. https://doi.org/10.1063/5.0004608
Grimme S, Bannwarth C, Dohm S et al (2017) Fully automated quantum-chemistry-based computation of spin-spin-coupled nuclear magnetic resonance spectra. Angew Chem Int Ed Engl 56:14763–14769. https://doi.org/10.1002/ANIE.201708266
Lu W, Zhang J, Huang W et al (2024) DynamicBind: predicting ligand-specific protein-ligand complex structure with a deep equivariant generative model. Nat Commun. https://doi.org/10.1038/S41467-024-45461-2
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/JCC.20084
Laskowski RA, Swindells MB (2011) LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786. https://doi.org/10.1021/CI200227U/ASSET/IMAGES/MEDIUM/CI-2011-00227U_0006.GIF
Acknowledgements
We appreciate the technical help in measuring QTOF-MS for Prof. Masaya Ohta (Fukuyama University, Hiroshima, Japan). In addition, we appreciate our collaborator Prof. Hiroshi Kamitakahara (Kyoto University, Kyoto, Japan), as a WP3 Leader of Japan-ASEAN Science, Technology and Innovation Platform (JASTIP).
Funding
This research was funded by the Japan Science and Technology Agency Strategic International Collaborative Research Program (SICORP, Japan), grant number JPMJSC15H1; JST MIRAI program, grant number JPMJMI22G1; JSPS KAKENHI, grant numbers 23K06189 and 24K02290; and the Japan Agency for Medicinal Research and Development (AMED), grant number JP23fk0108590.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Shoji Maehara and Moeka Kumamoto performed material preparation, data collection about compounds, and measurement and analysis of 3CL protease inhibition activity. Yuzoh Hieda synthesized ( +)-eupenoxide derivatives. Toshiyuki Hata performed computational analysis. Shogo Nakajima and Koichi Watashi evaluated the antiviral activity of SARS-CoV-2 using infected cells. Shoji Maehara wrote the first draft of the manuscript and all authors commented on previous versions of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest associated with this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Maehara, S., Kumamoto, M., Nakajima, S. et al. Potent SARS-CoV-2 3C-like protease inhibitor ( +)-eupenoxide-3,6-diketone (IC50: 0.048 μM) was synthesized based on ( +)-eupenoxide; lead from ( +)-eupenoxide analogs study by endophytic fermentation. J Nat Med 79, 357–370 (2025). https://doi.org/10.1007/s11418-024-01874-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11418-024-01874-3