Abstract
Objectives
To develop and validate a radiomics nomogram for timely predicting severe COVID-19 pneumonia.
Materials and methods
Three hundred and sixteen COVID-19 patients (246 non-severe and 70 severe) were retrospectively collected from two institutions and allocated to training, validation, and testing cohorts. Radiomics features were extracted from chest CT images. Radiomics signature was constructed based on reproducible features using the least absolute shrinkage and selection operator (LASSO) logistic regression algorithm with 5-fold cross-validation. Logistic regression modeling was employed to build different models based on quantitative CT features, radiomics signature, clinical factors, and/or the former combined features. Nomogram performance for severe COVID-19 prediction was assessed with respect to calibration, discrimination, and clinical usefulness.
Results
Sixteen selected features were used to build the radiomics signature. The CT-based radiomics model showed good calibration and discrimination in the training cohort (AUC, 0.9; 95% CI, 0.843–0.942), the validation cohort (AUC, 0.878; 95% CI, 0.796–0.958), and the testing cohort (AUC, 0.842; 95% CI, 0.761–0.922). The CT-based radiomics model showed better discrimination capability (all p < 0.05) compared with the clinical factors joint quantitative CT model (AUC, 0.781; 95% CI, 0.708–0.843) in the training cohort, the validation cohort (AUC, 0.814; 95% CI, 0.703–0.897), and the testing cohort (AUC, 0.696; 95% CI, 0.581–0.796). Decision curve analysis demonstrated that in terms of clinical usefulness, the radiomics model outperformed the clinical factors model and quantitative CT model alone.
Conclusions
The CT-based radiomics signature shows favorable predictive efficacy for severe COVID-19, which might assist clinicians in tailoring precise therapy.
Key Points
• Radiomics can be applied in CT images of COVID-19 and radiomics signature was an independent predictor of severe COVID-19.
• CT-based radiomics model can predict severe COVID-19 with satisfactory accuracy compared with subjective CT findings and clinical factors.
• Radiomics nomogram integrated with the radiomics signature, subjective CT findings, and clinical factors can achieve better severity prediction with improved diagnostic performance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corona Virus Disease 2019 (COVID-19) has spread out in the world, posing a critical threat to public health [1]. Initial report of the disease indicated that from November 16, 2019, to November 5, 2020, there were 47,596,852 confirmed cases of COVID-19, including 1,216,357 deaths, with a global fatality of approximately 2.56% [2].
According to the “Diagnosis and Treatment Program of Pneumonia of New Coronavirus Infection (Trial 7th Edition)” recommended by China’s National Health Commission, severe COVID-19 patients were more likely to develop poor clinical outcomes including acute respiratory distress syndrome (ARDS), acute cardiac/kidney injury, and multiple organ failure [3]. Current epidemiological data also suggests that the mortality rate of severe COVID-19 is about 20 times higher than that of non-severe COVID-19 [4]. Therefore, it is very crucial for early identification of severe cases, which prevent disease progression and reduce the mortality.
Computed tomography (CT) is widely used to diagnose, evaluate, and monitor COVID-19 pneumonia in high-risk areas [5]. However, the evaluation of these conventional CT features depends heavily on the radiologist’s experience and is non-objective. Furthermore, some scholars have tried using quantitative CT method to assess the severity of lung injury in COVID-19 pneumonia, such as visual CT severity score (CTSS) [6] and the CT lesion percentage (CTLP) [7] in each lung lobe, which were alternative ways to stratify COVID-2019 cases. The radiomics approach has drawn increased attention in recent years, which may help the detection, diagnosis, monitoring, and prognostic assessment of lung disease [8]. To our knowledge, there has been rarely radiomics study for the accurate prediction of the clinical severity of COVID-19 pneumonia to date.
Therefore, the aim of our study is to (1) investigate the role of radiomics features for predicting severe patients with COVID-19 pneumonia, and (2) examine whether the addition of quantitative CT characteristics and/or clinical factors can improve the performance of the predictive model.
Materials and methods
Patient population and groups
Our institutional review board approved this retrospective study, and the requirement for informed consent was waived.
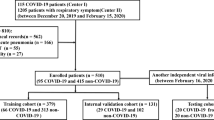
We searched the electronic database at two institutions ((I) Renmin Hospital of Wuhan University, (II) Central Hospital of Wuhan optical valley) and retrospectively reviewed records for patients between January 30, 2020, and April 30, 2020, and identified 449 patients infected with COVID-19. All patients with COVID-19 were proven using real-time reverse transcriptase–polymerase chain reaction. The whole pipeline of our study was shown in Fig. 1.
According to the “Diagnosis and Treatment Program of Pneumonia of New Coronavirus Infection (Trial 7th Edition)” recommended by China’s National Health Commission, all patients are classified as having minimal, ordinary, severe, and critical type [9]. In our study, ordinary cases were included in the non-severe disease (NSD) group, while severe and critical cases were merged into the severe disease (SD) group. All patients in the SD group should meet any of the following criteria: (1) respiratory rate ≥ 30 breaths per minute; (2) finger of oxygen saturation ≤ 93% in a resting state; (3) arterial oxygen tension (PaO2)/inspiratory oxygen fraction (FiO2) ≤ 300 mmHg (1 mmHg = 0.133 kPa); (4) respiratory failure occurred and mechanical ventilation required; (5) shock; (6) other organ failure needing intensive care unit (ICU) monitoring treatment.
Image acquisition and lesion segmentation
Non-enhanced chest CT scans of 316 patients were carried out from the lung apex to the lung base using multi-detector CT (MDCT) scanners (Brightspeed CT or Optima 680 CT, GE Healthcare) at the end of inspiration. Breath-hold training was carried out before each examination. Parameters for chest CT scanning were listed as follows: field of view (FOV), 36 cm; tube voltage, 120 kV; tube current adjusted automatically; noise index, 13; section thickness, 5 mm; slice interval, 5 mm; pitch, 1.375; collimation 64 × 0.625 mm; gantry rotation speed, 0.7 s; matrix, 512 × 512; the mediastinal window: window width of 200 HU with a window level of 35 HU, and the lung window: window width of 1500 HU with a window level of - 700 HU.
All images were segmented on the commercial segmentation software (Lung Intelligence Kit 2.1, LK 2.1, GE Healthcare) [10]. First, pre-processing was executed and included the following steps: resampling adjust the x-spacing, y-spacing, z-spacing size (spatial resolution = 1 mm × 1 mm × 1 mm). Gaussian filter with a standard deviation of 0.5 was applied for signal smoothing. Then, the lung was automatically segmented into five lobes; the volumes of interest (VOIs) were automatically contoured for each lobe. The segmentation results are manually corrected by a radiologist (Z.K., 2 years of experience in radiology) and then confirmed by another radiologist (F.Z., 5 years of experience in radiology). CTLP was defined as the volume of the lesions (including ground-glass opacity (GGO), consolidation, and reticulation) divided by the volume of the entire lung. CTSS was used to estimate pulmonary involvement of all abnormalities on the basis of the area involved [11]. Each of the five lung lobes was visually scored from 0 to 5 as follows: 0, no involvement; 1, less than 5% involvement; 2, less than 25% involvement; 3, 26–49% involvement; 4, 50–75% involvement; or 5, more than 75% involvement [12].
Collection of clinical data and evaluation of CT radiological features
Clinical data were recorded, including the following 10 characteristics: age, sex, duration of onset, comorbidity, clinical type, treatments, respiratory support strategies (RSS), ICU admission, length of ICU stay, and length of hospital stay. We also recorded other clinical parameters, such as oxygen saturation and temperature, but which were not used in the clinic’s model, because the former was one of the known diagnostic criteria for severe COVID-19, and the latter was a variable parameter due to uncertain medical history. Comorbidities included hypertension, diabetes, chronic obstructive pulmonary disease (COPD), chronic kidney disease (CKD), malignant tumors, and surgery history (on any part of the body in the last 10 years). The number of comorbidities was from 0 to 4; 0 means no complications, and 4 means there are 4 kinds of diseases. Treatments included antiviral therapy, antibiotic therapy, glucocorticoid therapy, immunoglobulin therapy, and Chinese medicine therapy. RSS included nasal catheter, high-flow nasal cannula oxygen therapy, non-invasive mechanical ventilation, invasive mechanical ventilation, and extracorporeal membrane oxygenation (ECMO).
All CT images were evaluated by 2 radiologists (Z.K. and L.L.) who were blinded to each subject’s clinical data. For disagreement between the two primary radiologist interpretations, a third experienced thoracic radiologist with 25 years of experience (Y.Z.) adjudicated a final decision. Ten CT radiological features were assessed, namely GGO, consolidation, reticular pattern, interlobular septal thickening, air bronchogram sign, lesion location, distribution, involved lobe, thickening of pleura, and pleural effusion.
Radiomics feature extraction
A total of 851 radiomics features were extracted from the VOIs segmented based on the L. K software, including first-order statistics parameters (n = 18), morphological parameters (n = 14), gray-level co-occurrence matrix (GLCM) parameters (n = 24), gray-level run length matrix (GLRLM) parameters (n = 16), gray-level size zone matrix (GLSZM) parameters (n = 16), gray-level dependence matrix (GLDM) parameters (n = 14), neighboring gray tone difference matrix (NGTDM) parameters (n = 5), and wavelet parameters (n = 744). All the features defined were in compliance with feature definitions as described by the Imaging Biomarker Standardization Initiative (IBSI) [13]. The detailed workflow of radiomics analysis can be found in Fig. 2. Intra- and interclass correlation coefficients (ICC) were used to assess the intra- and inter-observer reproducibility of radiomics feature extraction.
Radiomics features selection and radiomics signature construction
The outlier values were replaced by the median value of the particular variance vector once the values were beyond the range of the mean and standard deviation. And standardization was performed to scale the data in a specific interval. Spearman correlation, generalized linear model (GLM), and least absolute shrinkage and selection operator (LASSO) were used to reduce the redundancy or selection bias of the features, thereby removing a high correlation. A radiomics score (Rad-score) was calculated for each patient via a linear combination of selected features that were weighted by their respective coefficients.
Development of predictive models
The most significant features were investigated to construct radiomics model based on logistic regression. The likelihood ratio test with backward step-down selection was applied to the multivariate logistic regression model. We grouped the selected features into seven models—the radiomics model (radiomics features), the CTSS model (semi-quantitative CTSS), CTLP model (quantitative CTLP), the clinical model (clinical features), the integrated A model (CTSS + CTLP + clinical features), the integrated B model (clinical features + radiomics features), and the integrated C model (radiomics features + CTSS + CTLP + clinical features). The calibration curves were used to investigate the performance characteristics of the nomograms.
Statistical analysis
Statistical analyses were performed with the Institute of Precision Medicine Statistics (IPMs, version 1.1, GE Healthcare). The differences in all variables between NSD and SD groups were assessed using the Mann-Whitney U test or independent samples t test for continuous variables, and the chi-square test or Fisher’s exact test for categorical variables. Univariate analysis was used to estimate the relationship between clinical factors and the identification of the two subtypes. The performances of the seven models were assessed by area under the receiver operating characteristic curve (AUC), specificity, and sensitivity. The optimal cut-off points to predict the severity of COVID-19 were determined by Youden’s index. The DeLong test was used for pairwise comparisons among the seven models using the R software (v. 3.6.0; http://www.Rproject.org). A two-sided p < 0.05 was considered statistically significant throughout the study.
Results
Clinical and CT radiological features
The detailed distribution of clinical characteristics in the NSD and SD group was summarized in Table 1. Clinical factors (gender and duration of onset) were found not significantly different between the two groups in all cohorts, while the comorbidities and age were the only similarity between the two groups in the validation cohort.
Three hundred sixteen patients (NSD, 246 vs. SD, 70) from two hospitals were included in the study. The patients in both groups showed GGO with mainly peripheral distribution; there was no statistical difference (78% vs. 84%, p > 0.05). In the SD group, 45% of patients had consolidation, and 60% had multiple lung lobes involved, but only 35% and 50% in the NSD group (p < 0.05). More patients had pleural effusions in the SD group compared to that in the NSD group (54.28% vs. 7.72%, p < 0.05). More imaging details between the two groups in different hospitals are shown in Table 2.
Radiomics features selection and radiomics signature building
In total, 99 features with statistical significance (p < 0.05) between the NSD and SD groups were selected in the training dataset. A radiomics signature was further constructed based on sixteen features with respective non-zero coefficients selected from these 99 features. One first-order feature and fifteen wavelet-based features were used to represent the radiomics signature. Details of the procedure for the construction of the radiomics signature are showed in Fig. 3a, b. The lists of the selected features and their associated coefficients in the logistic regression model are illustrated in Fig. 3c. The intra-observer ICCs ranged from 0.822 to 0.957 and the inter-observer ICCs ranged from 0.769 to 0.936, indicating favorable intra- and inter-observer feature extraction reproducibility. Figure 4 shows representative images and lesion segmentation result of severe COVID-19
Texture feature selection using the least absolute shrinkage and selection operator (LASSO) binary logistic regression model. a LASSO coefficient profiles of the radiomics features. Vertical line was drawn at the value selected using 5-fold cross-validation in the ln(alpha) sequence, and 16 non-zero coefficients are indicated. b The tuning parameter λ selection in the LASSO model used 5-fold cross-validation via the minimum criteria. Mean square error was plotted vs. log (λ). The dotted vertical lines were drawn at the optimal values using the minimum criteria and the 1-SE criteria. c Multivariate logistic of the predictive radiomics features. OR, odds ratio
A case of confirmed severe COVID-19. A 62-year-old female presented with a 7-day history of fever and cough. First CT imaging revealed diffuse pure GGO with mainly peripheral distribution in the bilateral lobes (a). The area of the lesions was delineated on the axial, coronal, and reconstructed three-dimensional images (b, c, d). CTSS = total 10 scores, predicted probability for severe COVID-19 = 90.1%
The utility of severity prediction using developed radiomics signature
The developed initial CT-based radiomics signature model showed a favorable result in predicting the severity (NSD vs. SD) that produced an AUC of 0.9 in the training set (95% CI, 0.843 to 0.942), 0.878 in the internal validation set (95% CI, 0.796 to 0.958), and 0.842 in the testing set (95% CI, 0.761 to 0.922).
As shown in Fig. 5, predictive nomogram and correlation coefficients were built by combining Rad-score, age, comorbidity, L1-CTSS, L1-CTLP, R1-CTLP, and R2-CTLP. The predictive nomogram had the best differentiation ability of the severe cases with an AUC of 0.918 (95% CI, 0.864–0.956) in the training set, an AUC of 0.934 (95% CI, 0.848–0.979) in the validation set, and an AUC of 0.854 (95% CI, 0.762–0.92) in the testing set, as shown in Fig. 6a–c. The calibration curves of the nomogram also showed that the predictions agreed well with the observations in Fig. 6d–f.
The ROC curves of the seven prediction models that indicate severe COVID-19 cases in the training cohort (a), validation cohort (b), and testing cohort (c). Calibration curves of the combined nomogram in the training cohort (d), internal validation cohort (e), and testing cohort (f). Calibration curves depict the calibration of the nomogram in terms of agreement between the predicted risk and actual probability for severe COVID-19
Evaluation of models and comparison of predictive model performance
The diagnostic performance of each model is shown in Table 3 and the results of ROC curve analysis are shown in Fig. 6. In all cohorts, the integrated C model achieved the best performance and radiomics model outperformed clinic’s model for predicting severe COVID-19. Moreover, the AUC of the integrated B model was better than CTSS model and CTLP model (all p < 0.05). In the training set, although the integrated C model showed the highest AUC (0.918) among the seven predictive models, no statistical difference was found between any of the two models using the DeLong test (p = 0.832 for CTSS vs. CTLP; p = 0.165 for CTSS vs. clinics; p = 0.323 for CTLP vs. clinics). No statistical difference in AUC was also found between any of the two models (p = 0.09 for CTSS vs. integrated A; p = 0.137 for CTLP vs. integrated A; p = 0.751 for clinics vs. integrated A), but a statistical difference in AUC was found between the radiomics model and the integrated A model (p = 0.017), and between radiomics model and clinics model (p = 0.00075)
Treatment and prognosis in the patients with COVID-19
All 316 patients with COVID-19 received hospitalization, and their treatment information was recorded. Compared with the non-severe group, severe patients have more comorbidities and longer ICU and hospital length of stay (p < 0.05). Most patients received antibiotics (166 [52.53%]), antiviral treatment (276 [87.34%]), and glucocorticoids (178 [56.33%]). One hundred twenty-nine (40.82%) patients received intravenous infusions of immunoglobulin. Two-thirds of the patients had oxygen therapy. Treatment and prognosis information of all three cohorts are provided in Table 4.
Discussion
In this study, statistical analysis was performed in comparing clinical data, quantitative CT, and radiomics features between non-severe and severe COVID-19 patients. Two clinical features, four quantitative CT features, and sixteen radiomics features were identified to be significantly different between the two groups (p < 0.05). Furthermore, CT-based radiomics model had a significantly high area under the curve (AUC), sensitivity, and specificity to distinguish individuals with severe COVID-19 from non-severe COVID-19. The radiomics model alone was also validated with the independent dataset from the other institutions, suggesting the reliability and reproducibility of our new prediction model.
In general, the most common CT features for COVID-19 included GGO and consolidation involving the bilateral lungs in a peripheral distribution [14, 15]. In the early diagnostic stage, it is difficult for clinicians to judge whether it is COVID-19 pneumonia. Meanwhile, for non-severe and severe COVID-19, CT features are often highly overlapping and non-specific. Recently, some literatures reported that the proportion of lesions (GGO/consolidation) is different between NSD and SD patients [16,17,18]. Non-severe patients are more likely to have more GGO, while severe patients tend to have more consolidation in CT imaging. Although we have also observed this phenomenon, no statistical difference was found (p > 0.05). Therefore, it is also very difficult for the radiologist to judge whether it is severe pneumonia [19, 20].
Radiomics has great potential to capture useful medical information and to enhance the accuracy of clinical differential diagnosis. In this study, 851 candidate radiomics features were extracted from CT images and were reduced to only 16 potential predictors by using a LASSO regression model to develop the radiomics signatures. The selected radiomics features were divided into two types (first-order and wavelet features) and varied significantly between NSD and SD groups. These features reflect intrinsical information from the distribution of pixel intensity and the texture morphology that cannot be detected by radiologists [21]. For example, first-order features mainly reflect the internal texture of lesions. Wavelet features mainly reflect the change of time domain and frequency domain inside the lesion. Among the selected radiomics features in this study, Wavelet-HHL_glcm_Imc2 and Wavelet-LHL_glrlm_ShortRunLowGrayLevelEmphasis were the most significant and robust features associated with severe cases, which reflect lesion’s intensity and textural features within the high-intensity CT voxels.
Several studies have reported and analyzed the value of traditional CT characteristics and clinical features in the diagnosis of COVID-19. Chen et al [22] developed and validated a diagnostic model for COVID-19 based on CT imaging and clinical manifestations. Kang et al [23] introduced an AI-assisted model based on the radiological and clinical features to estimate the clinical progression to critical illness. In addition, in order to quantify the severity of COVID-19, CT quantification of pneumonia lesions is also used in clinical practice. The CTSS and CTLP are the two most common CT quantification methods, but the differences among observers are still inevitable [7, 16, 22]. In our model, L1-CTSS, L1-CTLP, R1-CTLP, and R2-CTLP were selected as independent risk factors for severe COVID-19 cases. The possible explanation is that the SARS-CoV-2 mainly invades the bilateral lower lobes in patients with COVID-19, so the difference in the distribution of lesions in the upper or middle lobe is often more statistically different among different patients with COVID-19.
The integrated C model composed of radiomics, quantitative CT, and clinical features had the highest AUC (0.92, 0.93, 0.84) in all three cohorts. The model with radiomics features alone reached an AUC of 0.9, 0.878, and 0.842, which is not inferior to the integrated B or C model (all p > 0.05). In other words, the addition of quantitative CT and clinical features to the integrated model did not increase the model’s efficacy, suggesting the strong efficacy of radiomics as a tool to predict the severity of COVID-19. Furthermore, the predictive power of the radiomics model also outperformed that of integrated A model (clinics + quantitative CT) (p < 0.05). In fact, radiomics features and clinical or quantitative CT features were highly correlated. This result was in accordance with that in other similar studies in gliomas [24], pancreatic neuroendocrine tumor (PNET) [23], lung adenocarcinoma [21], and breast cancer [25].
Several studies reported single or multiple risk factors of lung injury for severe COVID-19. Dong et al [26] found that underlying comorbidity, older age, higher LDH, and lower lymphocyte count were independent high-risk factors associated with COVID-19 progression in a multicenter study. Guan et al [27] reported that increasing age and comorbidity were associated with the disease. Zhou et al [28] showed that older age, high SOFA score, and D-dimer value are potential risk factors to identify patients with poor prognosis at an early stage. Chen et al [29] proposed the MuLBSTA score as an early warning indicator for predicting 90-day mortality in patients with COVID-19. Among the clinical features in our study, age and comorbidity were the most powerful factors to predict severe COVID-19. A possible reason is that poor immunosenescence of older patients with comorbidities could have higher levels of angiotensin-converting enzyme 2 in their alveoli [30], which was regarded as a receptor for novel coronaviruses [31].
Some limitations in this study should also be acknowledged. The biggest limitation is that no blood samples or some other clinical parameters are considered in the risk model. We believe that a reliable and robust multi-modality prediction model should be further developed to address this issue. In addition, the sample size was relatively small in order to build up a stable predicting model and the cases outside of Hubei province are not included. However, in this study, we tried to adopt a multicenter research method, including internal and external validation sets, subjective and objective CT evaluation, all of which ensure that the conclusions are reasonable. Furthermore, with the retrospective design, there may be potential biases in identifying and recruiting participants. Increasing a large number of different regional populations and imaging-pathologic control studies is needed in the future study.
In conclusion, initial CT-based radiomics features provide an excellent performance for the prediction of severe COVID-19. Sixteen features were significantly different between the two groups. This prediction model based on the radiomics features was developed and validated on the training, validation, and testing cohorts, which may have broad clinical applications.
Abbreviations
- COVID-19:
-
Corona Virus Disease 2019
- CTSS:
-
CT severity score
- CTLP:
-
CT lesion percentage
- DCA:
-
Decision curve analysis
- GGO:
-
Ground-glass opacity
- GLM:
-
Generalized linear model
- GLCM:
-
Gray-level co-occurrence matrix
- GLRLM:
-
Gray-level run length matrix
- ICC:
-
Intraclass correlation coefficients
- LASSO:
-
Least absolute shrinkage selection operator
- NGTDM:
-
Neighboring gray tone difference matrix
- NSD:
-
Non-severe disease
- ROC:
-
Receiver operating characteristic
- SD:
-
Severe disease
References
Zhu N, Zhang D, Wang W et al (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733
World Health Organization. Coronavirus disease (COVID-19) pandemic on 5 November 2020 [EB/OL]. https://www.whoint/emergencies/diseases/novel-coronavirus-2019. Accessed 5 Nov 2020
Huang C, Wang Y, Li X et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506
Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G (2020) Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. https://doi.org/10.1515/cclm-2020-0369
Xiong Y, Sun D, Liu Y et al (2020) Clinical and high-resolution CT features of the COVID-19 infection: comparison of the initial and follow-up changes. Invest Radiol 55:332–339
Ding X, Xu J, Zhou J, Long Q (2020) Chest CT findings of COVID-19 pneumonia by duration of symptoms. Eur J Radiol 127:109009
Shen C, Yu N, Cai S et al (2020) Quantitative computed tomography analysis for stratifying the severity of Coronavirus Disease 2019. J Pharm Anal. https://doi.org/10.1016/j.jpha.2020.03.004
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
The National HealthCommission of the People’s Republic of China. Diagnosis and treatment protocols of COVID-19 infection (trial version 7) [EB/OL]. http://www.nhc.gov.cn/yzygj/s7653p/202002/8334a8326dd94d329df351d7da8aefc2s.html. Accessed 3 Mar 2020
Wei W, Hu XW, Cheng Q, Zhao YM, Ge YQ (2020) Identification of common and severe COVID-19: the value of CT texture analysis and correlation with clinical characteristics. Eur Radiol 30:6788–6796
Pan F, Ye T, Sun P et al (2020) Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. https://doi.org/10.1148/radiol.2020200370:200370
Li K, Wu J, Wu F et al (2020) The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Invest Radiol 55:327–331
Zwanenburg A, Vallieres M, Abdalah MA et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295:328–338
Bao C, Liu X, Zhang H, Li Y, Liu J (2020) Coronavirus Disease 2019 (COVID-19) CT findings: a systematic review and meta-analysis. J Am Coll Radiol. https://doi.org/10.1016/j.jacr.2020.03.006
Zhou S, Wang Y, Zhu T, Xia L (2020) CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. AJR Am J Roentgenol. https://doi.org/10.2214/AJR.20.22975:1-8
Li K, Fang Y, Li W et al (2020) CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19). Eur Radiol 30:4407–4416
Pan F, Ye T, Sun P et al (2020) Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19). Radiology 295:715–721
Wang Y, Dong C, Hu Y et al (2020) Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology 296:E55–E64
Lyu P, Liu X, Zhang R, Shi L, Gao J (2020) The performance of chest CT in evaluating the clinical severity of COVID-19 pneumonia: identifying critical cases based on CT characteristics. Invest Radiol. https://doi.org/10.1097/RLI.0000000000000689
Wu J, Wu X, Zeng W et al (2020) Chest CT findings in patients with coronavirus disease 2019 and its relationship with clinical features. Invest Radiol 55:257–261
Song L, Zhu Z, Mao L et al (2020) Clinical, conventional CT and radiomic feature-based machine learning models for predicting ALK rearrangement status in lung adenocarcinoma patients. Front Oncol 10:369
Chen X, Tang Y, Mo Y et al (2020) A diagnostic model for coronavirus disease 2019 (COVID-19) based on radiological semantic and clinical features: a multi-center study. Eur Radiol. https://doi.org/10.1007/s00330-020-06829-2
Zhang K, Liu XH, Shen J et al (2020) Clinically applicable AI system for accurate diagnosis, quantitative measurements and prognosis of COVID-19 pneumonia using computed tomography. Cell. https://doi.org/10.1016/j.cell.2020.04.045
Wang Y, Wei W, Liu Z et al (2020) Predicting the type of tumor-related epilepsy in patients with low-grade gliomas: a radiomics study. Front Oncol 10:235
Liu Z, Li Z, Qu J et al (2019) Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res 25:3538–3547
Ji D, Zhang D, Xu J et al (2020) Prediction for progression risk in patients with COVID-19 pneumonia: the CALL score. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa414
Guan WJ, Ni ZY, Hu Y et al (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. https://doi.org/10.1056/NEJMoa2002032
Zhou F, Yu T, Du R et al (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054–1062
Guo L, Wei D, Zhang X et al (2019) Clinical features predicting mortality risk in patients with viral pneumonia: the MuLBSTA score. Front Microbiol 10:2752
Pera A, Campos C, Lopez N et al (2015) Immunosenescence: implications for response to infection and vaccination in older people. Maturitas 82:50–55
Reynolds HR, Adhikari S, Pulgarin C et al (2020) Renin-angiotensin-aldosterone system inhibitors and risk of Covid-19. N Engl J Med. https://doi.org/10.1056/NEJMoa2008975
Funding
This study has received funding from the National Natural Science Foundation of China (Nos. 81871332, 81601461), Novel Coronavirus Pneumonia Emergency Key Project of Science and Technology of Hubei Province (No. 2020FCA015), and the Fundamental Research Funds for the Central Universities (No. 2042020kfxg10).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Yunfei Zha.
Statistics and biometry
Statistician Huan Liu kindly provided all statistical work for this study.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Conflict of interest
One of the authors of this manuscript (Huan Liu) is an employee of GE Healthcare. The remaining authors declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, L., Wang, L., Zeng, F. et al. Development and multicenter validation of a CT-based radiomics signature for predicting severe COVID-19 pneumonia. Eur Radiol 31, 7901–7912 (2021). https://doi.org/10.1007/s00330-021-07727-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-07727-x