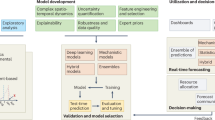
Abstract
The scenario of epidemic forecasting with inflection points and constant changes in population behavior is extremely critical. This way, there is a growing consensus in the process of modeling infectious diseases that forecasts should not only indicate a single point outcome but also quantify their own uncertainty. In this regard, our study investigated uncertainty quantification in the prediction of COVID-19 deaths, using Bayesian Deep Learning techniques and producing prediction intervals as an informational mechanism to address the unpredictability of external factors. Six distinct methods were employed: MC Dropout, Concrete Dropout, Spatial Dropout, MC-DropConnect, SG-HMC, and SWAG. Through regression heteroscedastic modeling, the approaches were tested and their results compared, particularly in periods with intermittent waves. The methodology initially emphasized building a robust database with an excellent representation of the problem. Subsequently, the experiments focused on forecasting daily intervals of the 7-day moving average, at the 28-day horizons, for the five most populous U.S. states, over a period of 1 year. Regarding the results, SWAG achieved the best results in the Log-Likelihood metric, while Spatial Dropout performed best in the MSE. The Concrete Dropout produced conservative predictive intervals with high coverage, above 85% using amplitudes between 13% and 34%. During the epidemiological wave, there was greater instability in the uncertainty estimation process, as evidenced by a reduction in the Log-Likelihood metric. The findings contribute to the maturity of a tangible and assertive perspective on uncertainty estimation in DL, facilitating decision-making in the dynamic environment of epidemic modeling.



Similar content being viewed by others
Data Availability
The database constructed, analyzed and processed during this work is available on the Harvard Dataverse repository at the following web address: https://doi.org/10.7910/DVN/PICBEA.
References
Baker RE, Mahmud AS, Miller IF, Rajeev M, Rasambainarivo F, Rice BL, Takahashi S, Tatem AJ, Wagner CE, Wang L-F, et al. Infectious disease in an era of global change. Nat Rev Microbiol. 2022;20(4):193–205. https://doi.org/10.1038/s41579-021-00639-z.
WHO: WHO Coronavirus Disease (COVID-19) Dashboard 2020; https://covid19.who.int/.
Chen Y, Liu F, Yu Q, Li T. Review of fractional epidemic models. Appl Math Model. 2021;97:281–307.
Bansal A, Jayant U. COVID-19 outbreak modelling using regression techniques. In: 2021 international conference on innovative practices in technology and management (ICIPTM). IEEE; 2021. pp. 113–118.
Khan MA, Khan R, Algarni F, Kumar I, Choudhary A, Srivastava A. Performance evaluation of regression models for COVID-19: a statistical and predictive perspective. Ain Shams Eng J. 2022;13(2): 101574.
Sahai AK, Rath N, Sood V, Singh MP. Arima modelling & forecasting of COVID-19 in top five affected countries. Diabetes Metab Syndr Clin Res Rev. 2020;14(5):1419–27.
ArunKumar K, Kalaga DV, Kumar CMS, Chilkoor G, Kawaji M, Brenza TM. Forecasting the dynamics of cumulative COVID-19 cases (confirmed, recovered and deaths) for top-16 countries using statistical machine learning models: auto-regressive integrated moving average (arima) and seasonal auto-regressive integrated moving average (sarima). Appl Soft Comput. 2021;103: 107161.
Germann TC, Smith MZ, Dauelsberg L, Fairchild G, Turton TL, Gorris ME, Ross CW, Ahrens JP, Hemphill DD, Manore C, et al. Using an agent-based model to assess k-12 school reopenings under different COVID-19 spread scenarios–United States, school year 2020/21. medRxiv, 2020;2020–10.
Errica F, Bacciu D, Micheli A. Graph mixture density networks. In: International conference on machine learning. PMLR; 2021. pp. 3025–3035.
Desai AN, Kraemer MU, Bhatia S, Cori A, Nouvellet P, Herringer M, Cohn EL, Carrion M, Brownstein JS, Madoff LC, et al. Real-time epidemic forecasting: challenges and opportunities. Health Secur. 2019;17(4):268–75.
Katris C. A time series-based statistical approach for outbreak spread forecasting: application of COVID-19 in Greece. Expert Syst Appl. 2021;166: 114077. https://doi.org/10.1016/j.eswa.2020.114077.
Du H, Dong E, Badr HS, Petrone ME, Grubaugh ND, Gardner LM. Incorporating variant frequencies data into short-term forecasting for COVID-19 cases and deaths in the USA: a deep learning approach. Ebiomedicine. 2023;89:104482.
Luo J. Forecasting COVID-19 pandemic: unknown unknowns and predictive monitoring. Technol Forecast Soc Chang. 2021;166: 120602.
Jewell NP, Lewnard JA, Jewell BL. Predictive mathematical models of the COVID-19 pandemic: underlying principles and value of projections. JAMA. 2020;323(19):1893–4.
Ioannidis JP, Cripps S, Tanner MA. Forecasting for COVID-19 has failed. Int J Forecast. 2022;38(2):423–38.
Shrestha DL, Solomatine DP. Machine learning approaches for estimation of prediction interval for the model output. Neural Netw. 2006;19(2):225–35. https://doi.org/10.1016/j.neunet.2006.01.012.
Der Kiureghian A, Ditlevsen O. Aleatory or epistemic? does it matter? Struct Saf. 2009;31(2):105–12.
Li Y, Chen J, Feng L. Dealing with uncertainty: a survey of theories and practices. IEEE Trans Knowl Data Eng. 2012;25(11):2463–82.
Helton JC. Treatment of uncertainty in performance assessments for complex systems. Risk Anal. 1994;14(4):483–511.
Meeker WQ, Hahn GJ, Escobar LA. Statistical intervals: a guide for practitioners and researchers vol. 541. John Wiley & Sons; 2017.
Pearce T, Brintrup A, Zaki M, Neely A. High-quality prediction intervals for deep learning: A distribution-free, ensembled approach. In: International conference on machine learning. PMLR; 2018. pp. 4075–4084.
Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian data analysis. Chapman and Hall/CRC; 2013.
Murphy KP. Machine learning: a probabilistic perspective. MIT press; 2012.
Gal Y, Ghahramani Z. Dropout as a bayesian approximation: Representing model uncertainty in deep learning. In: International conference on machine learning. PMLR; 2016. pp. 1050–1059.
Gal Y, Hron J, Kendall A. Concrete dropout. Adv Neural Inf Process Syst 2017;30
Mobiny A, Yuan P, Moulik SK, Garg N, Wu CC, Van Nguyen H. Dropconnect is effective in modeling uncertainty of Bayesian deep networks. Sci Rep. 2021;11(1):5458.
Chen T, Fox E, Guestrin C. Stochastic gradient hamiltonian monte carlo. In: International conference on machine learning. PMLR. 2014; pp. 1683–1691. https://proceedings.mlr.press/v32/cheni14.html.
Maddox WJ, Izmailov P, Garipov T, Vetrov DP, Wilson AG. A simple baseline for Bayesian uncertainty in deep learning. Adv Neural Inf Process Syst 2019;32
Gawlikowski J, Tassi CRN, Ali M, Lee J, Humt M, Feng J, Kruspe A, Triebel R, Jung P, Roscher R, et al. A survey of uncertainty in deep neural networks. Artif Intell Rev. 2023;56(Suppl 1):1513–89. https://doi.org/10.1007/s10462-023-10562-9.
Mukhoti J, Stenetorp P, Gal Y. On the importance of strong baselines in Bayesian deep learning. 2018; arXiv preprint arXiv:1811.09385.
Caldeira J, Nord B. Deeply uncertain: comparing methods of uncertainty quantification in deep learning algorithms. Mach Learn: Sci Technol. 2020;2(1): 015002.
Tran K, Neiswanger W, Yoon J, Zhang Q, Xing E, Ulissi ZW. Methods for comparing uncertainty quantifications for material property predictions. Mach Learn: Sci Technol. 2020;1(2): 025006.
Murayama T, Shimizu N, Fujita S, Wakamiya S, Aramaki E. Predicting regional influenza epidemics with uncertainty estimation using commuting data in Japan. PLoS ONE. 2021;16(4):0250417.
Zhu L, Laptev N. Deep and confident prediction for time series at uber. In: 2017 IEEE International Conference on Data Mining Workshops (ICDMW). IEEE. 2017; pp. 103–110.
Li Q, Yang Y, Wang W, Lee S, Xiao X, Gao X, Oztekin B, Fan C, Mostafavi A. Unraveling the dynamic importance of county-level features in trajectory of COVID-19. Sci Rep. 2021;11(1):13058.
CDC: Centers for Disease Control and Prevention 2022; https://www.cdc.gov/.
Ali N, Islam F. The effects of air pollution on COVID-19 infection and mortality—a review on recent evidence. Front Public Health. 2020;8: 580057.
McClymont H, Hu W. Weather variability and COVID-19 transmission: a review of recent research. Int J Environ Res Public Health. 2021;18(2):396.
Mollalo A, Rivera KM, Vahedi B. Artificial neural network modeling of novel coronavirus (COVID-19) incidence rates across the continental united states. Int J Environ Res Public Health. 2020;17(12):4204.
NCEI: National Centers for Environmental Information 2022; https://www.ncei.noaa.gov/.
US EPA: U.S. Environmental Protection Agency 2022; https://www.epa.gov/.
Google: Google COVID-19 Community Mobility Reports 2020; https://www.google.com/covid19/mobility/.
Apple: Mobility Trends Reports 2020; https://www.apple.com/covid19/mobility.
OxCGRT: Oxford COVID-19 Government Response Tracker 2022; https://www.bsg.ox.ac.uk/research/research-projects/covid-19-government-response-tracker.
CMS: Centers for Medicare & Medicaid Services (CMS) 2020; https://data.cms.gov/.
Liu K, Chen Y, Lin R, Han K. Clinical features of covid-19 in elderly patients: a comparison with young and middle-aged patients. J Infect. 2020;80(6):14–8.
Leung C. Risk factors for predicting mortality in elderly patients with COVID-19: a review of clinical data in China. Mech Ageing Dev. 2020;188: 111255.
CDC: Centers for Disease Control and Prevention. https://data.cdc.gov/Laboratory-Surveillance/SARS-CoV-2-Variant-Proportions/jr58-6ysp.
Ma KC. Genomic surveillance for sars-cov-2 variants: circulation of omicron lineages-United States, January 2022-May 2023. MMWR. Morb Mortal Weekly Rep. 2023 https://doi.org/10.15585/mmwr.mm7224a2
Blundell C, Cornebise J, Kavukcuoglu K, Wierstra D. Weight uncertainty in neural network. In: International conference on machine learning. PMLR; 2015. pp. 1613–1622.
Amini A, Soleimany A, Karaman S, Rus D. Spatial uncertainty sampling for end-to-end control. 2018; arXiv preprint arXiv:1805.04829
Krueger D, Huang C-W, Islam R, Turner R, Lacoste A, Courville A. Bayesian hypernetworks. 2017; arXiv preprint arXiv:1710.04759
Kendall A, Gal Y. What uncertainties do we need in Bayesian deep learning for computer vision? Adv Neural Inf Process Syst 2017;30
Eisenstein M. Infection forecasts powered by big data. Nature. 2018;555(7697):2–2.
Khosravi A, Nahavandi S, Creighton D. A prediction interval-based approach to determine optimal structures of neural network metamodels. Expert Syst Appl. 2010;37(3):2377–87.
Khosravi A, Nahavandi S, Creighton D, Atiya AF. Lower upper bound estimation method for construction of neural network-based prediction intervals. IEEE Trans Neural Networks. 2010;22(3):337–46.
Cramer EY, Ray EL, Lopez VK, Bracher J, Brennen A, Castro Rivadeneira AJ, Gerding A, Gneiting T, House KH, Huang Y, et al. Evaluation of individual and ensemble probabilistic forecasts of COVID-19 mortality in the united states. Proc Natl Acad Sci. 2022;119(15):2113561119.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no Conflict of interest.
Ethical Approval
No animal or human experimentation was involved in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gonçalves, A.V., de Araújo, G.M. & Júnior, E.M.d.S. Evaluation of Bayesian Deep Learning Methods for Quantifying Uncertainties in Forecasting Deaths from COVID-19. SN COMPUT. SCI. 6, 276 (2025). https://doi.org/10.1007/s42979-025-03829-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-025-03829-1