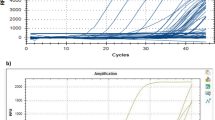
Abstract
Background The emergence and rapid spread of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), poses a significant threat to human health and public safety. While next-generation sequencing (NGS) is capable of detecting and tracking new COVID-19 variants for disease diagnosis and prevention, its high cost and time-consuming nature limit its widespread use. In this study, our aim was to develop a highly adaptable and accurate RT-PCR method for identifying the Delta or BA.1 variants in inactivated COVID-19 vaccine. We devised three two-plex RT-PCR methods targeting specific mutation sites: S: Δ156–157, S: N211-, L212I, and S: Δ142–144, Y145D. The RT-PCR method targeting the S: Δ156–157 mutation site was able to distinguish the Delta variant from other COVID-19 virus strains, while the RT-PCR methods targeting the S: N211-, L212I or S: Δ142–144, Y145D mutation sites were able to distinguish the BA.1 variant from other COVID-19 virus strains. We separately validated these three two-plex RT-PCR methods, and the results demonstrated good linearity, repeatability, reproducibility, and specificity for each method. Moreover, all three methods can be applied in the production of SARS-CoV-2 variant inactivated vaccines, enabling the identification of Delta or BA.1 variants in virus cultures as well as in inactivated vaccine stocks. This study presents a systematic approach to identify COVID-19 variants using multiple RT-PCR methods. We successfully developed three two-plex RT-PCR methods that can identify Delta and BA.1 variants based on specific mutation sites, and we completed the validation of these three methods.





Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Data availability
No datasets were generated or analysed during the current study.
References
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R et al (2020) A novel coronavirus from patients with Pneumonia in China, 2019. N Engl J Med 382:727–733. https://doi.org/10.1056/NEJMoa2001017
Ciotti M, Angeletti S, Minieri M, Giovannetti M, Benvenuto D, Pascarella S, Sagnelli C, Bianchi M, Bernardini S, Ciccozzi M (2019) COVID-19 outbreak: an overview. Chemotherapy 64:215–223. https://doi.org/10.1159/000507423
Scovino AM, Dahab EC, Vieira GF, Freire-de-Lima L, Freire-de-Lima CG (2022) Morrot, A. SARS-CoV-2’s variants of concern: a brief characterization. Front Immunol 13:834098. https://doi.org/10.3389/fimmu.2022.834098
Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Peacock SJ et al (2021) SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. https://doi.org/10.1038/s41579-021-00573-0
Zhang Y, Zhang H, Zhang W (2022) SARS-CoV-2 variants, immune escape, and countermeasures. Front Med 16:196–207. https://doi.org/10.1007/s11684-021-0906-x
Zhang Y, Tan W, Lou Z, Huang B, Zhou W, Zhao Y, Zhang J, Liang H, Li N, Zhu X et al (2022) Immunogenicity evaluating of the multivalent COVID-19 inactivated vaccine against the SARS-CoV-2 variants. Vaccines 10:956. https://doi.org/10.3390/vaccines10060956
Wang R, Chen J, Hozumi Y, Yin C, Wei GW (2022) Emerging vaccine-breakthrough SARS-CoV-2 variants. ACS Infect Dis 8:546
Willett BJ, Grove J, MacLean OA, Wilkie C, De Lorenzo G, Furnon W, Cantoni D, Scott S, Logan N, Ashraf S et al (2022) SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol 7:1161–1179. https://doi.org/10.1038/s41564-022-01143-7
Carreño JM, Alshammary H, Tcheou J, Singh G, Raskin AJ, Kawabata H, Sominsky LA, Clark JJ, Adelsberg DC, Bielak DA et al (2022) Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 602:682–688. https://doi.org/10.1038/s41586-022-04399-5
Bouhaddou M, Reuschl AK, Polacco BJ, Thorne LG, Ummadi MR, Ye C, Rosales R, Pelin A, Batra J, Jang GM et al (2023) SARS-CoV-2 variants evolve convergent strategies to remodel the host response. Cell 186:4597–4614.e26. https://doi.org/10.1016/j.cell.2023.08.026
Torjesen I (2021) Covid-19: Delta variant is now UK’s most dominant strain and spreading through schools. BMJ 373:n1445. https://doi.org/10.1136/bmj.n1445
Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J et al (2021) Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596:276–280. https://doi.org/10.1038/s41586-021-03777-9
Karim SSA, Karim QA (2021) Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet 398:2126–2128. https://doi.org/10.1016/S0140-6736(21)02758-6
John G, Sahajpal NS, Mondal AK, Ananth S, Williams C, Chaubey A, Rojiani AM, Kolhe R (2021) Next-Generation sequencing (NGS) in COVID-19: a tool for SARS-CoV-2 diagnosis, monitoring new strains and phylodynamic modeling in molecular epidemiology. Curr Issues Mol Biol 43:845–867. https://doi.org/10.3390/cimb43020061
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt M L et al (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro. Surveill 25:2000045. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
Lu X, Sakthivel SK, Wang L, Lynch B, Dollard SM (2021) Enhanced throughput of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) real-time RT-PCR panel by assay multiplexing and specimen pooling. J Virol Methods 293:114149. https://doi.org/10.1016/j.jviromet.2021.114149
Wang H, Zhang Y, Huang B, Deng W, Quan Y, Wang W, Xu W, Zhao Y, Li N, Zhang J et al (2020) Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell 182:713–721.e9. https://doi.org/10.1016/j.cell.2020.06.008
Xia S, Zhang Y, Wang Y, Wang H, Yang Y, Gao GF, Tan W, Wu G, Xu M, Lou Z et al (2021) Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect Dis 21:39–51. https://doi.org/10.1016/S1473-3099(20)30831-8
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML et al (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Erratum. In: Euro. Surveill 25:2000045. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
Spiess K, Gunalan V, Marving E, Nielsen SH, Jørgensen MGP, Fomsgaard AS, Nielsen L, Alfaro-Núñez A, Karst SM, Mortensen S et al (2023) Rapid and flexible RT-qPCR surveillance platforms to detect sars-cov-2 mutations. Microbiol Spectr 11:e0359122. https://doi.org/10.1128/spectrum.03591-22
Nörz D, Grunwald M, Tang HT, Olearo F, Günther T, Robitaille A, Fischer N, Grundhoff A, Aepfelbacher M, Pfefferle S et al (2021) Rapid automated screening for SARS-CoV-2 B.1.617 lineage variants (Delta/Kappa) through a versatile toolset of qpcr-based SNP detection. Diagnostics 11:1818. https://doi.org/10.3390/diagnostics11101818
Gazali FM, Nuhamunada M, Nabilla R, Supriyati E, Hakim MS, Arguni E, Daniwijaya EW, Nuryastuti T, Haryana SM, Wibawa T et al (2021) Detection of SARS-CoV-2 spike protein D614G mutation by qPCR-HRM analysis. Heliyon 7:e07936. https://doi.org/10.1016/j.heliyon.2021.e07936
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
Y.Z. and Y.Z. are the corresponding author and coordinate the strains and experimental sites used in this study. Z.W. and Y.H. conceived and designed this study. Z.W. prepared the manuscript. Z.W. Y.H. Z.H. and Y.G. performed all experiments. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethical approval
Not applicable.
Competing interests
The authors declare no competing interests
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Z., He, Y., He, Z. et al. Development of highly adaptable RT-PCR methods for identifying Delta and BA.1 variants in inactivated COVID-19 vaccines. Mol Biol Rep 51, 892 (2024). https://doi.org/10.1007/s11033-024-09799-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09799-6