Abstract
Introduction
The worldwide COVID-19 pandemic is a serious problem for people around the world, especially for healthcare professionals. Currently, two COVID testing techniques are in vogue which includes RT-PCR and Rapid Antigen test. The RT-PCR is an expensive detection technique, and it takes approximately 24 h to test and process the sample in order to formulate the results. The detection of the virus at an earliest with higher accuracy and at optimal price are the prerequisites that need to be addressed by any detection technique proposed by the researchers. The cheaper and quicker method is known as Rapid Antigen test or rapid test. However, these tests are less reliable, with an accuracy rate in some cases as low as 50%. This requires the development of detection methods that target large populations and reduce the rate of detection errors by alleviating the burden on the health care system.
Objective
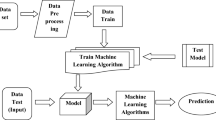
In this article, we have developed several machine learning models, namely (logistic regression, SVM, K-NN, and NB) utilizing various algorithms to predict COVID-19 based on the exhibited symptoms. The primary aim is to enable quick and accurate testing of individuals affected by the coronavirus. RESULTS: Through our work, we have achieved diverse accuracies (91.38%, 54.77%, 95.49%, and 91.57%) while utilizing different algorithms. Consequently, we have implemented our model with the linear SVM, which has an accuracy of 95.49%, an F-score of 92.08%, precision of 93.01%, and a recall of 91.16% for real-time detection.








Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Alakus TB, Turkoglu I. Comparison of deep learning approaches to predict COVID-19 infection. Chaos Solit Fractals. 2020;140:110120.
Asif S, Wenhui Y, Jin H, Jinha S. Classification of COVID-19 from chest X-ray images using deep convolutional neural network. In: 2020 IEEE 6th international conference on computer and communications (ICCC). Chengdu, China; 2020. p. 426–33. IEEE.
Asif S, Zhao M, Tang F, Zhu Y. A deep learning-based framework for detecting COVID-19 patients using chest X-rays. Multimedia Syst. 2022;28(4):1495–513.
Altay O, Ulas M. Prediction of the autism spectrum disorder diagnosis with linear discriminant analysis classifier and K-nearest neighbor in children. In: 2018 6th International Symposium on Digital Forensic and Security (ISDFS). Antalya, Turkey; 2018. p. 1–4. IEEE.
Bullock J, Luccioni A, Pham KH, Lam CSN, Luengo-Oroz M. Mapping the landscape of artificial intelligence applications against COVID-19. J Artif Intell Res. 2020;69. AI Access Foundation.
Brunese L, Martinelli F, Mercaldo F, Santone A. Machine learning for coronavirus COVID-19 detection from chest x-rays. Procedia Comput Sci. 2020;176:2212–21.
Chavez S, Long B, Koyfman A, Liang SY. Coronavirus disease (COVID-19): a primer for emergency physicians. Am J Emerg Med. 2020;44:220–229. [google scholar] [crossref] [pubmed].
Delizo JPD, Abisado MB, Trinos MID. Philippine twitter sentiments during COVID-19 pandemic using multinomial Naïve-Bayes. Int J Adv Trends Comput Sci Eng. 2020;9:408–12.
Guo H, Zhou Y, Liu X, Tan J. The impact of the COVID-19 epidemic on the utilization of emergency dental services. J Dent Sci. 2020;15:564–567. [google scholar] [crossref] [pubmed].
Hosmer RXSDW, Lemeshow S. Applied Logistic Regression. Toronto, Canada: John Wiley & Sons; 2013.
Kassani SH, Kassasni PH, Wesolowski MJ, Schneider KA, Deters R. Automatic detection of coronavirus disease (COVID-19) in x-ray and ct images: a machine learning-based approach. arXiv. 2020;41:867–79.
Khanday AMUD, Rabani ST, Khan QR, et al. Machine learning based approaches for detecting COVID-19 using clinical text data. Int J Inf Technol. 2020;12:731–9.
Roberts M, Driggs D, Thorpe M, et al. Cornrnon pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Lntell. 2021;3:199–217.
Routledge R. Bayes’s theorem, 17 February 2018. https://www.britannica.com/topic/Bayess-theorem
Sarwar A, Ali M, Manhas J, et al. Diagnosis of diabetes type-ii using hybrid machine learning based ensemble model. Int J Inf Technol. 2018;12:419–28. Springer.
Shah C, Jivani, A. Comparison of data mining classification algorithms for breast cancer prediction. In Proceedings of the 4th ICCCNT 2013, Tiruchengode, India, 4–6 July 2013.
Villavicencio CN, Macrohon JJE, Inbaraj X, Jeng J-H, Hsieh J-G. Twitter sentiment analysis towards COVID-19 vaccines using Naive Bayes. Information 2021;12. MDPI.
WHO. Middle East respiratory syndrome coronavirus (mers-cov). 2021a. Available online: https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers#tab=tab_1. Accessed 3 Dec 2021.
WHO. Naming the coronavirus disease (COVID-2019) and the virus that causes it. 2021b. Available online:https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-thevirus-that-causes-it. Accessed 3 Dec 2021.
Zoabi Y, Deri-Rozov S, Shomron N. Machine learning-based prediction of COVID-19 diagnosis based on symptoms. NPJ Digit Med. 2021;4:1-5, Springer.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hameed, J., Khan, U.R., Noor, S. et al. SARS-COV-2 (COVID-19) detection application via machine learning: comparative analysis and performance evaluation. Res. Biomed. Eng. 39, 925–935 (2023). https://doi.org/10.1007/s42600-023-00316-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42600-023-00316-5