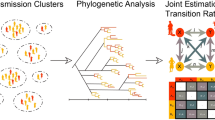
Abstract
Precise identification of HIV transmission among populations is a key step in public health responses. However, the HIV transmission network is usually difficult to determine. HIV molecular networks can be determined by phylogenetic approach, genetic distance-based approach, and a combination of both approaches. These approaches are increasingly used to identify transmission networks among populations, reconstruct the history of HIV spread, monitor the dynamics of HIV transmission, guide targeted intervention on key subpopulations, and assess the effects of interventions. Simulation and retrospective studies have demonstrated that these molecular network-based interventions are more cost-effective than random or traditional interventions. However, we still need to address several challenges to improve the practice of molecular network-guided targeting interventions to finally end the HIV epidemic. The data remain limited or difficult to obtain, and more automatic real-time tools are required. In addition, molecular and social networks must be combined, and technical parameters and ethnic issues warrant further studies.
Article PDF
Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Avoid common mistakes on your manuscript.
References
McMahon JH, Medland N. 90-90-90: how do we get there? Lancet HIV 2014; 1(1): e20–e11
Kazanjian P. UNAIDS 90-90-90 campaign to end the AIDS epidemic in historic perspective. Milbank Q 2017; 95(2): 408–439
Machuca R, Jørgensen LB, Theilade P, Nielsen C. Molecular investigation of transmission of human immunodeficiency virus type 1 in a criminal case. Clin Diagn Lab Immunol 2001; 8(5): 884–890
Leitner T, Escanilla D, Franzén C, Uhlén M, Albert J. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc Natl Acad Sci USA 1996; 93(20): 10864–10869
Trask SA, Derdeyn CA, Fideli U, Chen Y, Meleth S, Kasolo F, Musonda R, Hunter E, Gao F, Allen S, Hahn BH. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J Virol 2002; 76(1): 397–405
Smith DM, May SJ, Tweeten S, Drumright L, Pacold ME, Kosakovsky Pond SL, Pesano RL, Lie YS, Richman DD, Frost SD, Woelk CH, Little SJ. A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS 2009; 23(2): 225–232
Aldous JL, Pond SK, Poon A, Jain S, Qin H, Kahn JS, Kitahata M, Rodriguez B, Dennis AM, Boswell SL, Haubrich R, Smith DM. Characterizing HIV transmission networks across the United States. Clin Infect Dis 2012; 55(8): 1135–1143
Little SJ, Kosakovsky Pond SL, Anderson CM, Young JA, Wertheim JO, Mehta SR, May S, Smith DM. Using HIV networks to inform real time prevention interventions. PLoS One 2014; 9(6): e98443
Volz EM, Le Vu S, Ratmann O, Tostevin A, Dunn D, Orkin C, O’Shea S, Delpech V, Brown A, Gill N, Fraser C; UK HIV Drug Resistance Database. Molecular epidemiology of HIV-1 subtype B reveals heterogeneous transmission risk: implications for intervention and control. J Infect Dis 2018; 217(10): 1522–1529
Oster AM, France AM, Mermin J. Molecular epidemiology and the transformation of HIV prevention. JAMA 2018; 319(16): 1657–1658
National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention; Division of HIV/AIDS. Prevention Detecting and Responding to HIV Transmission Clusters: A Guide for Health Department. 2018. https://www.cdc.gov/hiv/pdf/funding/announcements/ps18-1802/CDC-HIV-PS18-1802-AttachmentE-Detecting-Investigating-and-Responding-to-HIV-Transmission-Clusters.pdf
Fauci AS, Redfield RR, Sigounas G, Weahkee MD, Giroir BP. Ending the HIV epidemic: a plan for the United States. JAMA 2019; 321(9): 844–845
Hué S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS 2004; 18(5): 719–728
Yebra G, Hodcroft EB, Ragonnet-Cronin ML, Pillay D, Brown AJ; PANGEA_HIV Consortium; ICONIC Project. Using nearly full-genome HIV sequence data improves phylogeny reconstruction in a simulated epidemic. Sci Rep 2016; 6(1): 39489
Yerly S, Vora S, Rizzardi P, Chave JP, Vernazza PL, Flepp M, Telenti A, Battegay M, Veuthey AL, Bru JP, Rickenbach M, Hirschel B, Perrin L; Swiss HIV Cohort Study. Acute HIV infection: impact on the spread of HIV and transmission of drug resistance. AIDS 2001; 15(17): 2287–2292
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4(4): 406–425
Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 2012; 29(9): 2157–2167
Baele G, Li WL, Drummond AJ, Suchard MA, Lemey P. Accurate model selection of relaxed molecular clocks in bayesian phylogenetics. Mol Biol Evol 2013; 30(2): 239–243
Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, Matschiner M, Mendes FK, Müller NF, Ogilvie HA, du Plessis L, Popinga A, Rambaut A, Rasmussen D, Siveroni I, Suchard MA, Wu CH, Xie D, Zhang C, Stadler T, Drummond AJ. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol 2019; 15(4): e1006650
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 2018; 4(1): vey016
Dennis AM, Hué S, Billock R, Levintow S, Sebastian J, Miller WC, Eron JJ. Human immunodeficiency virus type 1 phylodynamics to detect and characterize active transmission clusters in North Carolina. J Infect Dis 2019 Apr 27. [Epub ahead of print] doi: https://doi.org/10.1093/infdis/jiz176
Wilkinson E, Junqueira DM, Lessells R, Engelbrecht S, van Zyl G, de Oliveira T, Salemi M. The effect of interventions on the transmission and spread of HIV in South Africa: a phylodynamic analysis. Sci Rep 2019; 9(1): 2640
Poon AF. Impacts and shortcomings of genetic clustering methods for infectious disease outbreaks. Virus Evol 2016; 2(2): vew031
Aldous JL, Pond SK, Poon A, Jain S, Qin H, Kahn JS, Kitahata M, Rodriguez B, Dennis AM, Boswell SL, Haubrich R, Smith DM. Characterizing HIV transmission networks across the United States. Clin Infect Dis 2012; 55(8): 1135–1143
Chin BS, Chaillon A, Mehta SR, Wertheim JO, Kim G, Shin HS, Smith DM. Molecular epidemiology identifies HIV transmission networks associated with younger age and heterosexual exposure among Korean individuals. J Med Virol 2016; 88(10): 1832–1835
Wang X, Wu Y, Mao L, Xia W, Zhang W, Dai L, Mehta SR, Wertheim JO, Dong X, Zhang T, Wu H, Smith DM. Targeting HIV prevention based on molecular epidemiology among deeply sampled subnetworks of men who have sex with men. Clin Infect Dis 2015; 61(9): 1462–1468
Rose R, Lamers SL, Dollar JJ, Grabowski MK, Hodcroft EB, Ragonnet-Cronin M, Wertheim JO, Redd AD, German D, Laeyendecker O. Identifying transmission clusters with cluster picker and HIV-TRACE. AIDS Res Hum Retroviruses 2017; 33(3): 211–218
Prosperi MC, Ciccozzi M, Fanti I, Saladini F, Pecorari M, Borghi V, Di Giambenedetto S, Bruzzone B, Capetti A, Vivarelli A, Rusconi S, Re MC, Gismondo MR, Sighinolfi L, Gray RR, Salemi M, Zazzi M, De Luca A; ARCA collaborative group. A novel methodology for large-scale phylogeny partition. Nat Commun 2011; 2(1): 321
Ragonnet-Cronin M, Hodcroft E, Hué S, Fearnhill E, Delpech V, Brown AJ, Lycett S; UK HIV Drug Resistance Database. Automated analysis of phylogenetic clusters. BMC Bioinformatics 2013; 14(1): 317
Hassan AS, Pybus OG, Sanders EJ, Albert J, Esbjörnsson J. Defining HIV-1 transmission clusters based on sequence data. AIDS 2017; 31(9): 1211–1222
Paraskevis D, Magiorkinis E, Magiorkinis G, Kiosses VG, Lemey P, Vandamme AM, Rambaut A, Hatzakis A. Phylogenetic reconstruction of a known HIV-1 CRF04_cpx transmission network using maximum likelihood and Bayesian methods. J Mol Evol 2004; 59(5): 709–717
Esbjörnsson J, Mild M, Audelin A, Fonager J, Skar H, Bruun Jørgensen L, Liitsola K, Björkman P, Bratt G, Gisslén M, Sönnerborg A, Nielsen C; SPREAD/ESAR Programme, Medstrand P, Albert J. HIV-1 transmission between MSM and heterosexuals, and increasing proportions of circulating recombinant forms in the Nordic Countries. Virus Evol 2016; 2(1): vew010
Bruhn CA, Audelin AM, Helleberg M, Bjorn-Mortensen K, Obel N, Gerstoft J, Nielsen C, Melbye M, Medstrand P, Gilbert MT, Esbjrnsson J. The origin and emergence of an HIV-1 epidemic: from introduction to endemicity. AIDS 2014; 28(7): 1031–1040
Jovanović L, Šiljić M, Ćirković V, Salemović D, Pešić-Pavlović I, Todorović M, Ranin J, Jevtović D, Stanojević M. Exploring evolutionary and transmission dynamics of HIV epidemic in Serbia: bridging socio-demographic with phylogenetic approach. Front Microbiol, 2019, 10: 287
Bbosa N, Ssemwanga D, Nsubuga RN, Salazar-Gonzalez JF, Salazar MG, Nanyonjo M, Kuteesa M, Seeley J, Kiwanuka N, Bagaya BS, Yebra G, Leigh-Brown A, Kaleebu P. Phylogeography of HIV-1 suggests that Ugandan fishing communities are a sink for, not a source of, virus from general populations. Sci Rep 2019; 9(1): 1051
Ragonnet-Cronin M, Jackson C, Bradley-Stewart A, Aitken C, McAuley A, Palmateer N, Gunson R, Goldberg D, Milosevic C, Leigh Brown AJ. Recent and rapid transmission of HIV among people who inject drugs in Scotland revealed through phylogenetic analysis. J Infect Dis 2018; 217(12): 1875–1882
Paraskevis D, Beloukas A, Stasinos K, Pantazis N, de Mendoza C, Bannert N, Meyer L, Zangerle R, Gill J, Prins M, d’Arminio Montforte A, Kran AB, Porter K, Touloumi G; CASCADE collaboration of EuroCoord. HIV-1 molecular transmission clusters in nine European countries and Canada: association with demographic and clinical factors. BMC Med 2019; 17(1): 4
de Oliveira T, Kharsany AB, Gräf T, Cawood C, Khanyile D, Grobler A, Puren A, Madurai S, Baxter C, Karim QA, Karim SS. Transmission networks and risk of HIV infection in KwaZulu-Natal, South Africa: a community-wide phylogenetic study. Lancet HIV 2017; 4(1): e41–e50
Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pépin J, Posada D, Peeters M, Pybus OG, Lemey P. The early spread and epidemic ignition of HIV-1 in human populations. Science 2014; 346(6205): 56–61
Waruru A, Achia TNO, Tobias JL, Nganga J, Mwangi M, Wamicwe J, Zielinski-Gutierrez E, Oluoch T, Muthama E, Tylleskär T. Finding hidden HIV clusters to support geographic-oriented HIV interventions in Kenya. J Acquir Immune Defic Syndr 2018; 78(2): 144–154
Butt Z, Grady S, Wilkins M, Hamilton E, Todem D, Gardiner J, Saeed M. Spatial epidemiology of HIV-hepatitis co-infection in the State of Michigan: a cohort study. Infect Dis (Lond) 2015; 47(12): 852–861
Stecher M, Hoenigl M, Eis-Hbinger AM, Lehmann C, Fätkenheuer G, Wasmuth JC, Knops E, Vehreschild JJ, Mehta S, Chaillon A. Hotspots of transmission driving the local human immunodeficiency virus epidemic in the Cologne-Bonn Region, Germany. Clin Infect Dis 2019; 68(9): 1539–1546
Chaillon A, Essat A, Frange P, Smith DM, Delaugerre C, Barin F, Ghosn J, Pialoux G, Robineau O, Rouzioux C, Goujard C, Meyer L, Chaix ML; on behalf the ANRS PRIMO Cohort Study. Spatiotemporal dynamics of HIV-1 transmission in France (1999–2014) and impact of targeted prevention strategies. Retrovirology 2017; 14(1): 15
Wertheim JO, Leigh Brown AJ, Hepler NL, Mehta SR, Richman DD, Smith DM, Kosakovsky Pond SL. The global transmission network of HIV-1. J Infect Dis 2014; 209(2): 304–313
Fisher M, Pao D, Brown AE, Sudarshi D, Gill ON, Cane P, Buckton AJ, Parry JV, Johnson AM, Sabin C, Pillay D. Determinants of HIV-1 transmission in men who have sex with men: a combined clinical, epidemiological and phylogenetic approach. AIDS 2010; 24(11): 1739–1747
Yirrell DL, Pickering H, Palmarini G, Hamilton L, Rutemberwa A, Biryahwaho B, Whitworth J, Brown AJ. Molecular epidemiological analysis of HIV in sexual networks in Uganda. AIDS 1998; 12(3): 285–290
Ng KT, Ng KY, Chen JH, Ng OT, Kamarulzaman A, Tee KK. HIV-1 transmission networks among men who have sex with men in Asia. Clin Infect Dis 2014; 59(6): 910–911
Mehta SR, Wertheim JO, Brouwer KC, Wagner KD, Chaillon A, Strathdee S, Patterson TL, Rangel MG, Vargas M, Murrell B, Garfein R, Little SJ, Smith DM. HIV transmission networks in the San Diego-Tijuana Border Region. EBioMedicine 2015; 2(10): 1456–1463
Oster AM, Wertheim JO, Hernandez AL, Ocfemia MC, Saduvala N, Hall HI. Using molecular HIV surveillance data to understand transmission between subpopulations in the United States. J Acquir Immune Defic Syndr 2015; 70(4): 444–451
Ragonnet-Cronin M, Hu YW, Morris SR, Sheng Z, Poortinga K, Wertheim JO. HIV transmission networks among transgender women in Los Angeles County, CA, USA: a phylogenetic analysis of surveillance data. Lancet HIV 2019; 6(3): e164–e172
Li X, Liu H, Liu L, Feng Y, Kalish ML, Ho SYW, Shao Y. Tracing the epidemic history of HIV-1 CRF01_AE clusters using near-complete genome sequences. Sci Rep 2017; 7(1): 4024
Li Z, He X, Wang Z, Xing H, Li F, Yang Y, Wang Q, Takebe Y, Shao Y. Tracing the origin and history of HIV-1 subtype B′ epidemic by near full-length genome analyses. AIDS 2012; 26(7): 877–884
Ye J, Lu H, Su X, Xin R, Bai L, Xu K, Yu S, Feng X, Yan H, He X, Zeng Y. Phylogenetic and temporal dynamics of human immunodeficiency virus type 1B in China: four types of B strains circulate in China. AIDS Res Hum Retroviruses 2014; 30(9): 920–926
Meng Z, Xin R, Zhong P, Zhang C, Abubakar YF, Li J, Liu W, Zhang X, Xu J. A new migration map of HIV-1 CRF07_BC in China: analysis of sequences from 12 provinces over a decade. PLoS One 2012; 7(12): e52373
Chen X, Ye M, Pang W, Smith DM, Zhang C, Zheng YT. First appearance of HIV-1 CRF07_BC and CRF08_BC outside China. AIDS Res Hum Retroviruses 2017; 33(1): 74–76
Han X, Takebe Y, Zhang W, An M, Zhao B, Hu Q, Xu J, Wu H, Wu J, Lu L, Chen X, Liang S, Wang Z, Yan H, Fu J, Cai W, Zhuang M, Liao C, Shang H. A large-scale survey of CRF55_01B from men-who-have-sex-with-men in China: implying the evolutionary history and public health impact. Sci Rep 2015; 5(1): 18147
Han X, An M, Zhang M, Zhao B, Wu H, Liang S, Chen X, Zhuang M, Yan H, Fu J, Lu L, Cai W, Takebe Y, Shang H. Identification of 3 distinct HIV-1 founding strains responsible for expanding epidemic among men who have sex with men in 9 Chinese cities. J Acquir Immune Defic Syndr 2013; 64(1): 16–24
An M, Han X, Xu J, Chu Z, Jia M, Wu H, Lu L, Takebe Y, Shang H. Reconstituting the epidemic history of HIV strain CRF01_AE among men who have sex with men (MSM) in Liaoning, northeastern China: implications for the expanding epidemic among MSM in China. J Virol 2012; 86(22): 12402–12406
Li X, Xue Y, Lin Y, Gai J, Zhang L, Cheng H, Ning Z, Zhou L, Zhu K, Vanham G, Kang L, Wang Y, Zhuang M, Pan Q, Zhong P. Evolutionary dynamics and complicated genetic transmission network patterns of HIV-1 CRF01_AE among MSM in Shanghai, China. Sci Rep 2016; 6(1): 34729
Zhu Z, Hu Y, Xing W, Guo M, Zhao R, Han S, Wu B. Identifying symptom clusters among people living with HIV on antiretroviral therapy in China: a network analysis. J Pain Symptom Manage 2019; 57(3): 617–626
Zhang Z, Dai L, Jiang Y, Feng K, Liu L, Xia W, Yu F, Yao J, Xing W, Sun L, Zhang T, Wu H, Su B, Qiu M. Transmission network characteristics based on env and gag sequences from MSM during acute HIV-1 infection in Beijing, China. Arch Virol 2017; 162(11): 3329–3338
Li X, Zhu K, Xue Y, Wei F, Gao R, Duerr R, Fang K, Li W, Song Y, Du G, Yan W, Musa TH, Ge Y, Ji Y, Zhong P, Wei P. Multiple introductions and onward transmission of HIV-1 subtype B strains in Shanghai, China. J Infect 2017; 75(2): 160–168
Chen M, Ma Y, Chen H, Dai J, Dong L, Yang C, Li Y, Luo H, Zhang R, Jin X, Yang L, Cheung AKL, Jia M, Song Z. HIV-1 genetic transmission networks among men who have sex with men in Kunming, China. PLoS One 2018; 13(4): e0196548
Shao Y. AIDS molecular network research and AIDS precision prevention and control. 2018 National Conference on HIV and Hepatitis C Prevention and Treatment. April 19, 2018
Wang X, He X, Zhong P, Liu Y, Gui T, Jia D, Li H, Wu J, Yan J, Kang D, Han Y, Li T, Yang R, Han X, Chen L, Zhao J, Xing H, Liang S, He J, Yan Y, Xue Y, Zhang J, Zhuang X, Liang S, Bao Z, Li T, Zhuang D, Liu S, Han J, Jia L, Li J, Li L. Phylodynamics of major CRF01_AE epidemic clusters circulating in mainland of China. Sci Rep 2017; 7(1): 6330
Valverde EE, Oster AM, Xu S, Wertheim JO, Hernandez AL. HIV transmission dynamics among foreign-born persons in the United States. J Acquir Immune Defic Syndr 2017; 76(5): 445–452
Campbell EM, Jia H, Shankar A, Hanson D, Luo W, Masciotra S, Owen SM, Oster AM, Galang RR, Spiller MW, Blosser SJ, Chapman E, Roseberry JC, Gentry J, Pontones P, Duwve J, Peyrani P, Kagan RM, Whitcomb JM, Peters PJ, Heneine W, Brooks JT, Switzer WM. Detailed transmission network analysis of a large opiate-driven outbreak of HIV infection in the United States. J Infect Dis 2017; 216(9): 1053–1062
Poon AFY, Gustafson R, Daly P, Zerr L, Demlow SE, Wong J, Woods CK, Hogg RS, Krajden M, Moore D, Kendall P, Montaner JSG, Harrigan PR. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study. Lancet HIV 2016; 3(5): e231–e238
Pasquale DK, Doherty IA, Sampson LA, Hué S, Leone PA, Sebastian J, Ledford SL, Eron JJ, Miller WC, Dennis AM. Leveraging phylogenetics to understand HIV transmission and partner notification networks. J Acquir Immune Defic Syndr 2018; 78(4): 367–375
Avila D, Keiser O, Egger M, Kouyos R, Böni J, Yerly S, Klimkait T, Vernazza PL, Aubert V, Rauch A, Bonhoeffer S, Günthard HF, Stadler T, Spycher BD; Swiss HIV Cohort Study. Social meets molecular: combining phylogenetic and latent class analyses to understand HIV-1 transmission in Switzerland. Am J Epidemiol 2014; 179(12): 1514–1525
Wertheim JO, Kosakovsky Pond SL, Forgione LA, Mehta SR, Murrell B, Shah S, Smith DM, Scheffler K, Torian LV. Social and genetic networks of HIV-1 transmission in New York City. PLoS Pathog 2017; 13(1): e1006000
Kostaki EG, Nikolopoulos GK, Pavlitina E, Williams L, Magiorkinis G, Schneider J, Skaathun B, Morgan E, Psichogiou M, Daikos GL, Sypsa V, Smyrnov P, Korobchuk A, Malliori M, Hatzakis A, Friedman SR, Paraskevis D. Molecular analysis of human immunodeficiency virus type 1 (HIV-1)-infected individuals in a network-based intervention (Transmission Reduction Intervention Project): phylogenetics identify HIV-1-infected individuals with social links. J Infect Dis 2018; 218(5): 707–715
Leigh Brown AJ, Lycett SJ, Weinert L, Hughes GJ, Fearnhill E, Dunn DT; UK HIV Drug Resistance Collaboration. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis 2011; 204(9): 1463–1469
France AM, Panneer N, Ocfemia CB, Saduvala N, Campbell E, Switzer WM, Wertheim J, Oster AM. Rapidly growing HIV transmission clusters in the Unites States, 2013–2016. 2018 Conference on Retroviruses and Opportunistic Infections. March 4–7, 2018
Wertheim JO, Oster AM, Switzer WM, Zhang C, Panneer N, Campbell E, Saduvala N, Johnson JA, Heneine W. Natural selection favoring more transmissible HIV detected in United States molecular transmission network. Nat Commun 2019; 10(1): 5788
Peters PJ, Pontones P, Hoover KW, Patel MR, Galang RR, Shields J, Blosser SJ, Spiller MW, Combs B, Switzer WM, Conrad C, Gentry J, Khudyakov Y, Waterhouse D, Owen SM, Chapman E, Roseberry JC, McCants V, Weidle PJ, Broz D, Samandari T, Mermin J, Walthall J, Brooks JT, Duwve JM; Indiana HIV Outbreak Investigation Team. HIV infection linked to injection use of oxymorphone in Indiana, 2014–2015. N Engl J Med 2016; 375(3): 229–239
Monterosso A, Minnerly S, Goings S, Morris A, France AM, Dasgupta S, Oster AM, Fanning M. Identifying and investigating a rapidly growing HIV transmission cluster in Texas. Conference on Retroviruses and Opportunistic Infections. March 8, 2017. Seattle, Washington
Shang H, Xu J, Han X, Spero Li J, Arledge KC, Zhang L. HIV prevention: bring safe sex to China. Nature 2012; 485(7400): 576–577
Wertheim JO, Murrell B, Mehta SR, Forgione LA, Kosakovsky Pond SL, Smith DM, Torian LV. Growth of HIV-1 molecular transmission clusters in New York City. J Infect Dis 2018; 218(12): 1943–1953
Mehta SR, Murrell B, Anderson CM, Kosakovsky Pond SL, Wertheim JO, Young JA, Freitas L, Richman DD, Mathews WC, Scheffler K, Little SJ, Smith DM. Using HIV sequence and epidemiologic data to assess the effect of self-referral testing for acute HIV infection on incident diagnoses in San Diego, California. Clin Infect Dis 2016; 63(1): 101–107
Heesterbeek H, Anderson RM, Andreasen V, Bansal S, De Angelis D, Dye C, Eames KT, Edmunds WJ, Frost SD, Funk S, Hollingsworth TD, House T, Isham V, Klepac P, Lessler J, Lloyd-Smith JO, Metcalf CJ, Mollison D, Pellis L, Pulliam JR, Roberts MG, Viboud C; Isaac Newton Institute IDD Collaboration. Modeling infectious disease dynamics in the complex landscape of global health. Science 2015; 347(6227): aaa4339
Stadler T, Kouyos R, von Wyl V, Yerly S, Böni J, Bürgisser P, Klimkait T, Joos B, Rieder P, Xie D, Günthard HF, Drummond AJ, Bonhoeffer S; Swiss HIV Cohort Study. Estimating the basic reproductive number from viral sequence data. Mol Biol Evol 2012; 29(1): 347–357
Turk T, Bachmann N, Kadelka C, Büni J, Yerly S, Aubert V, Klimkait T, Battegay M, Bernasconi E, Calmy A, Cavassini M, Furrer H, Hoffmann M, Günthard HF, Kouyos RD, Aubert V, Battegay M, Bernasconi E, Böni J, Braun DL, Bucher HC, Calmy A, Cavassini M, Ciuffi A, Dollenmaier G, Egger M, Elzi L, Fehr J, Fellay J, Furrer H, Fux CA, Günthard HF, Haerry D, Hasse B, Hirsch HH, Hoffmann M, Hösli I, Kahlert C, Kaiser L, Keiser O, Klimkait T, Kouyos RD, Kovari H, Ledergerber B, Martinetti G, Martinez de Tejada B, Marzolini C, Metzner KJ, Müller N, Nicca D, Pantaleo G, Paioni P, Rauch A, Rudin C, Scherrer AU, Schmid P, Speck R, Stöckle M, Tarr P, Trkola A, Vernazza P, Wandeler G, Weber R, Yerly S. Assessing the danger of self-sustained HIV epidemics in heterosexuals by population based phylogenetic cluster analysis. eLife, 2017, 6: e28721
France AM, Oster AM. The promise and complexities of detecting and monitoring HIV transmission clusters. J Infect Dis 2019 Apr 27. [Epub ahead of print] doi: https://doi.org/10.1093/infdis/jiz177
Novitsky V, Moyo S, Lei Q, DeGruttola V, Essex M. Impact of sampling density on the extent of HIV clustering. AIDS Res Hum Retroviruses 2014; 30(12): 1226–1235
Chaillon A, Delaugerre C, Brenner B, Armero A, Capitant C, Nere ML, Leturque N, Pialoux G, Cua E, Tremblay C, Smith DM, Goujard C, Meyer L, Molina JM, Chaix ML. In-depth sampling of high-risk populations to characterize HIV transmission epidemics among young MSM using PrEP in France and Quebec. Open Forum Infect Dis 2019; 6(3): ofz080
Kosakovsky Pond SL, Weaver S, Leigh Brown AJ, Wertheim JO. HIV-TRACE (Transmission Cluster Engine): a tool for large scale molecular epidemiology of HIV-1 and other rapidly evolving pathogens. Mol Biol Evol 2018; 35(7): 1812–1819
Dennis AM, Pasquale DK, Billock R, Beagle S, Mobley V, Cope A, Kuruc J, Sebastian J, Walworth C, Leone PA. Integration of contact tracing and phylogenetics in an investigation of acute HIV infection. Sex Transm Dis 2018; 45(4): 222–228
Fitzmaurice AG, Linley L, Zhang C, Watson M, France AM, Oster AM. Novel method for rapid detection of spatiotemporal HIV clusters potentially warranting intervention. Emerg Infect Dis 2019; 25(5): 988–991
Hightower GK, May SJ, Pérez-Santiago J, Pacold ME, Wagner GA, Little SJ, Richman DD, Mehta SR, Smith DM, Pond SL. HIV-1 clade B pol evolution following primary infection. PLoS One 2013; 8(6): e68188
Hemelaar J, Elangovan R, Yun J, Dickson-Tetteh L, Fleminger I, Kirtley S, Williams B, Gouws-Williams E, Ghys PD; WHO-UNAIDS Network for HIV Isolation Characterisation. Global and regional molecular epidemiology of HIV-1, 1990–2015: a systematic review, global survey, and trend analysis. Lancet Infect Dis 2019; 19(2): 143–155
Bbosa N, Kaleebu P, Ssemwanga D. HIV subtype diversity worldwide. Curr Opin HIV AIDS 2019; 14(3): 153–160
Fabeni L, Alteri C, Berno G, Scutari R, Orchi N, De Carli G, Bertoli A, Carioti L, Gori C, Forbici F, Salpini R, Vergori A, Gagliardini R, Cicalini S, Mondi A, Pinnetti C, Mazzuti L, Turriziani O, Colafigli M, Borghi V, Montella F, Pennica A, Lichtner M, Girardi E, Andreoni M, Mussini C, Antinori A, Ceccherini-Silberstein F, Perno CF, Santoro MM4; SENDIH Study group. Characterisation of HIV-1 molecular transmission clusters among newly diagnosed individuals infected with non-B subtypes in Italy. Sex Transm Infect 2019; 95 (8): 619–625
Bon I, Ciccozzi M, Zehender G, Biagetti C, Verrucchi G, Lai A, Lo Presti A, Gibellini D, Re MC. HIV-1 subtype C transmission network: the phylogenetic reconstruction strongly supports the epidemiological data. J Clin Virol 2010; 48(3): 212–214
Parczewski M, Leszczyszyn-Pynka M, Bander D, Urbanska A, Boroć-Kaczmarska A. HIV-1 subtype D infections among Caucasians from Northwestern Poland—phylogenetic and clinical analysis. PLoS One 2012; 7(2): e31674
Mehta SR, Schairer C, Little S. Ethical issues in HIV phylogenetics and molecular epidemiology. Curr Opin HIV AIDS 2019; 14(3): 221–226
Prosperi MC, De Luca A, Di Giambenedetto S, Bracciale L, Fabbiani M, Cauda R, Salemi M. The threshold bootstrap clustering: a new approach to find families or transmission clusters within molecular quasispecies. PLoS One 2010; 5(10): e13619
Vrbik I, Stephens DA, Roger M, Brenner BG. The Gap Procedure: for the identification of phylogenetic clusters in HIV-1 sequence data. BMC Bioinformatics 2015; 16(1): 355
Kosakovsky Pond SL, Weaver S, Leigh Brown AJ, Wertheim JO. HIV-TRACE (TRAnsmission Cluster Engine): a tool for large scale molecular epidemiology of HIV-1 and other rapidly evolving pathogens. Mol Biol Evol 2018; 35(7): 1812–1819
Hué S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS 2004; 18(5): 719–728
Acknowledgements
This work was supported in part by the Mega-Projects of the National Science Research for the 13th Five-Year Plan (No. 2017ZX10201101), Innovation Team Development Program of the Ministry of Education (No. IRT_16R70), the National Natural Science Foundation of China (No. 81871637), and Central Public-interest Scientific Institution Basal Research Fund (No. 2018PT-31042).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Xiaoxu Han, Bin Zhao, Minghui An, Ping Zhong, and Hong Shang declare no conflicts of interest. This manuscript is a review article and does not entail a research protocol requiring approval by the relevant institutional review board or ethics committee.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Han, X., Zhao, B., An, M. et al. Molecular network-based intervention brings us closer to ending the HIV pandemic. Front. Med. 14, 136–148 (2020). https://doi.org/10.1007/s11684-020-0756-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11684-020-0756-y