Abstract
The Omicron (B.1.1.529) variant of SARS-CoV-2 emerged in November 2021 and is rapidly spreading among the human population1. Although recent reports reveal that the Omicron variant robustly escapes vaccine-associated and therapeutic neutralization antibodies2,3,4,5,6,7,8,9,10, the pathogenicity of the virus remains unknown. Here we show that the replication of Omicron is substantially attenuated in human Calu3 and Caco2 cells. Further mechanistic investigations reveal that Omicron is inefficient in its use of transmembrane serine protease 2 (TMPRSS2) compared with wild-type SARS-CoV-2 (HKU-001a) and previous variants, which may explain its reduced replication in Calu3 and Caco2 cells. The replication of Omicron is markedly attenuated in both the upper and lower respiratory tracts of infected K18-hACE2 mice compared with that of the wild-type strain and Delta (B.1.617.2) variant, resulting in its substantially ameliorated lung pathology. Compared with wild-type SARS-CoV-2 and the Alpha (B.1.1.7), Beta (1.351) and Delta variants, infection by Omicron causes the lowest reduction in body weight and the lowest mortality rate. Overall, our study demonstrates that the replication and pathogenicity of the Omicron variant of SARS-CoV-2 in mice is attenuated compared with the wild-type strain and other variants.
Similar content being viewed by others
Main
COVID-19 is caused by SARS-CoV-2 (refs. 11,12). Since its emergence in 2019, SARS-CoV-2 continues to evolve by acquiring mutations that increase its transmissibility, modulate its pathogenicity or confer resistance to neutralization antibodies. A new SARS-CoV-2 variant (PANGO lineage B.1.1.529) was reported on 24 November 2021 from Botswana and South Africa. Within two days, the Technical Advisory Group on Virus Evolution of the World Health Organization (WHO) classified B.1.1.529 as the fifth variant of concern (VOC), and designated it the Omicron variant1.
Omicron is characterized by an unusually large number of mutations in its spike protein, including 30 amino acid substitutions, three short deletions and one insertion, compared with the original strain of SARS-CoV-2. Among these changes, 15 of the amino acid substitutions are located in the receptor-binding domain with additional amino acid substitutions located in or near to the furin-like cleavage site at the S1/S2 junction and in S2 that may modulate host protease cleavage by furin and transmembrane serine proteases. The abundant mutations at these regions hinted that Omicron may escape neutralizing antibodies in the sera of convalescent or vaccinated individuals, and have modified capacity in cell entry, replication and pathogenesis.
Preliminary reports suggest that the Omicron variant is highly transmissible, even in fully vaccinated individuals and individuals who have received a booster dose13,14,15. These results are congruous with recent findings that the Omicron variant is markedly resistant to neutralization by sera not only from convalescent patients, but also from individuals vaccinated with many widely used COVID-19 vaccines2,3,4,5,6,7. Moreover, the neutralization ability of the monoclonal antibodies in clinical use against COVID-19 is abolished or severely impaired against the Omicron variant8,9,10. In this regard, the Omicron variant represents a critical threat to the pandemic control and disease treatment of the COVID-19 pandemic.
The number of infections caused Omicron is growing rapidly, with a propensity to replace the Delta variant as the predominant circulating SARS-CoV-2 variant. In contrast to its ability to escape antibody neutralization, whether or not the Omicron variant causes a milder or more severe disease remains unknown. Here we evaluated the virological features and pathogenesis of Omicron in vitro and in vivo, and compared the results with that of SARS-CoV-2 wild type (WT) and other VOCs. Our results demonstrate that the replication ability of Omicron is substantially attenuated both in vitro and in vivo compared with SARS-CoV-2 WT and other VOCs, which is explained by its lowered efficiency in using TMPRSS2. In the K18-hACE2 mouse model, the Omicron variant replicated significantly less efficiently than SARS-CoV-2 WT and Delta in both nasal turbinates and lungs, and induced substantially attenuated lung pathology. Finally, by comparing body weight loss and survival, our results suggest that the Omicron variant has attenuated pathogenicity compared with SARS-CoV-2 WT and previous VOCs, including Alpha, Beta and Delta.
The attenuated replication of Omicron
The Delta variant replaced all other VOCs and was the predominant circulating SARS-CoV-2 variant since mid-2021. Infection caused by Omicron has been increasing rapidly since late November 2021, and it may become the next predominant circulating SARS-CoV-2 variant. We first compared the replication efficiency of Omicron with that of SARS-CoV-2 WT, Alpha, Beta and Delta. We included Omicron(R346K) in the comparison, which contains a R346K substitution in the spike protein that represents 8.5% (956 out of 11,242) of the total Omicron variant sequences deposited into GISAID database as of 19 December 2021. By measuring the subgenomic envelope (sgE) gene that represents replication intermediates, our results suggested that the replication of Omicron was severely attenuated in Calu3 human lung epithelial cells. Area under the curve (AUC) quantification of the sgE gene over a period of 48 h suggested that Omicron and Omicron(R346K) replicated 3.4-fold and 4.2-fold less efficiently compared with the SARS-CoV-2 WT (Fig. 1a). By contrast, the Alpha, Beta and Delta variants replicated to similar or higher levels compared with the SARS-CoV-2 WT in Calu3 cells (Fig. 1a). We next measured the amount of infectious virus particles produced from Calu3 cells infected with the SARS-CoV-2 WT and variants. Consistent with the sgE gene results, we demonstrated that, although the Alpha, Beta and Delta variants produced similar or higher levels of infectious virus particles between 8–48 h post-infection (h.p.i.), the Omicron variant produced significantly less infectious virus particles compared with SARS-CoV-2 WT in Calu3 cells (Fig. 1b). At 48 h.p.i., the infectious virus titres of Omicron and Omicron(R346K) were 501.4-fold (P = 0.0163) and 114.7-fold (P = 0.0168) lower than that of SARS-CoV-2 WT, respectively (Fig. 1b). In parallel, we evaluated the replication of Omicron in Caco2 human intestinal epithelial cells. Similar to the results in Calu3 cells, the replication of the Omicron variant was substantially lower compared with that of SARS-CoV-2 WT, Alpha, Beta and Delta (Extended Data Fig. 1a). At 48 h.p.i., the infectious virus titres of Omicron and Omicron(R346K) were 407.2-fold (P = 0.0123) and 238.1-fold (P = 0.0124) lower compared with that of SARS-CoV-2 WT, respectively (Fig. 1c). Interestingly, in VeroE6 cells, the replication of the Omicron variant was reduced to a much lesser extent compared with that of SARS-CoV-2 WT, and was at a comparable level to the Delta variant (Fig. 1d and Extended Data Fig. 1b). We next evaluated the cytopathic effect of the Omicron variant in VeroE6-TMPRSS2 cells. Our results demonstrated that both Omicron and Omicron(R346K) were less cytopathic than SARS-CoV-2 WT, as evidenced by the significantly higher cell viability between 12–36 h.p.i. (Fig. 1e). Overall, these findings indicate that Omicron replicates less efficiently than SARS-CoV-2 WT, Alpha, Beta and Delta, and induces a comparatively lower level of cell damage in the infected cells.
a–d, Cells were challenged with SARS-CoV-2 WT, Alpha, Beta, Delta, Omicron or Omicron(R346K) at a multiplicity of infection (m.o.i.) of 0.5 (for Calu3) or 0.1 (for Caco2 and VeroE6). a, Cell lysates were collected at the designated time points for quantifying the subgenomic RNA of the envelope gene (sgE). n = 7. The robustness of sgE production was quantified by AUC analysis. b–d, Infectious viral particles were titrated using median tissue culture infectious dose (TCID50) assays in Calu3 (b), Caco2 (c) and VeroE6 cells (d). n = 4. e, The cell viability of VeroE6-TMPRSS2 cells infected with SARS-CoV-2 WT and other variants at 0.1 m.o.i. was quantified at the designated time points. n = 6. Data are mean ± s.d. from the indicated number of biological repeats. Statistical significance was determined using one-way analysis of variance (ANOVA) (a), two-tailed Student’s t-tests (b–d) and two-way ANOVA (e); where multiple comparisons were performed among different groups, P values were adjusted using Tukey’s multiple-comparison test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; NS, not significant. Data were obtained from three independent experiments. Each data point represents one biological replicate.
Mechanism of the attenuated replication of Omicron
Our live virus infection assays demonstrate that the replication of Omicron is markedly attenuated in Calu3 and Caco2 cells compared with the replication of SARS-CoV-2 WT and the other variants in these cells, whereas, in VeroE6 cells, Omicron replication is only modestly decreased. Our results are consistent with a recent preprint showing a decrease in pseudovirus entry of the Omicron variant in Calu3 and Caco2 cells, but not in Vero, HEK293T or A549-ACE2 cells16. Previous reports demonstrated that SARS-CoV-2 enters Calu3 and Caco2 cells through the plasma membrane entry pathway mediated by TMPRSS2, whereas the virus enters TMPRSS2-deficient cells (Vero, VeroE6, HEK293T and A549) through the endocytic pathway mediated by cathepsin L or other endosomal proteases17,18,19. Combining previous knowledge and current evidence from us and others, we postulate that the spike mutations of the Omicron variant might result in the reduced ability of the virus to use TMPRSS2, severely impairing its entry and replication in Calu3 and Caco2 cells, but not in VeroE6 cells. To evaluate this possibility, we assessed the entry of pseudoviruses carrying the spike protein of SARS-CoV-2 WT or the Alpha, Beta, Delta or Omicron variants side by side in HEK293T cells transfected with ACE2 with or without additional TMPRSS2 overexpression. Our results demonstrate that TMPRSS2 overexpression increased SARS-CoV-2 WT, Alpha, Beta and Delta pseudovirus entry in HEK293T-ACE2 cells by 11.3-fold, 12.6- fold, 11.7- fold and 16-fold, respectively (Fig. 2a). By contrast, TMPRSS2 overexpression increased Omicron pseudovirus entry by only 4.3-fold, which was significantly lower compared with that of SARS-CoV-2 WT or the other variants (P < 0.0001) (Fig. 2a). To further validate this result, we compared pseudovirus entry in VeroE6 and VeroE6-TMPRSS2 cells side by side. Our results demonstrated that TMPRSS2 expression in the VeroE6-TMPRSS2 stable cell line increased pseudovirus entry for SARS-CoV-2 WT, Alpha, Beta and Delta by 16.1-fold (P < 0.0001), 7.9-fold (P = 0.0005), 32.1-fold (P < 0.0001) and 48.7-fold (P < 0.0001), respectively. By contrast, the Omicron pseudovirus was less sensitive to TMPRSS2 expression and its entry was marginally increased by only 1.3-fold (not significant) in VeroE6-TMPRSS2 cells compared with that in VeroE6 cells (Fig. 2b). We next evaluated pseudovirus entry in VeroE6-TMPRSS2 cells in the presence of camostat, a potent TMPRSS2 inhibitor. Our data show that the inhibitory effect of camostat on Omicron pseudovirus entry was significantly reduced compared with that of the WT, Alpha, Beta or Delta pseudovirus (Fig. 2c). Camostat was similarly less potent in inhibiting Omicron and Omicron(R346K) replication compared with WT and other variants in VeroE6-TMPRSS2 cells (Fig. 2d). Consistent with these results, we detected decreased cleavage of Omicron spike compared with SARS-CoV-2 WT and Delta spike in the infected VeroE6-TMPRSS2 cells (Extended Data Fig. 2). Collectively, our results indicate that the reduced replication ability of the Omicron variant in cells that the virus enters predominantly through the plasma membrane entry pathway, including Calu3, Caco2 and VeroE6-TMPRSS2, is caused by its reduced spike cleavage, leading to inefficient recognition by TMPRSS2.
a, HEK293T cells were transfected with ACE2 or co-transfected with ACE2 and TMPRSS2, followed by transduction with pseudoviruses carrying the spike protein of SARS-CoV-2 WT, Alpha, Beta, Delta or Omicron at 24 h.p.i. Pseudovirus entry was quantified by measuring the luciferase signal. n = 9 (Delta) and n = 10 (WT, Alpha, Beta and Omicron). Fold changes in the luciferase signal were normalized to the mean luciferase readouts of cells with only ACE2 overexpression. b, VeroE6 and VeroE6-TMPRSS2 cells were transduced with pseudoviruses carrying the spike protein of SARS-CoV-2 WT, Alpha, Beta, Delta or Omicron. Pseudovirus entry was quantified by measuring the luciferase signal. n = 6. Fold changes in the luciferase signal were normalized to the mean luciferase readouts of VeroE6 cells. c, d, VeroE6-TMPRSS2 cells were pretreated with 1 μM, 25 μM or 50 μM camostat or DMSO for 2 h followed by transduction with pseudoviruses carrying the spike protein of SARS-CoV-2 WT, Alpha, Beta, Delta or Omicron (c) or challenged with the indicated authentic SARS-CoV-2 variants at 0.1 m.o.i. (d). Pseudovirus entry was quantified by measuring the luciferase signal of the cell lysates at 24 h.p.i. n = 6. The amount of viral subgenomic envelope RNA in collected cell lysates at 24 h.p.i. was determined using quantitative PCR with reverse transcription (RT–qPCR). n = 7 (Delta, Omicron and Omicron(R346K) at 1 µM, 25 µM and 50 µM) and n = 6 (all other samples). Data are mean ± s.d. from the indicated number of biological replicates. Statistical significance was determined using two-way ANOVA (a–d); where multiple comparisons were performed among different groups, P values were adjusted using Tukey’s multiple-comparison test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; NS, not significant. Data were obtained from three independent experiments. Each data point represents one biological replicate.
Replication and pathogenesis of Omicron in vivo
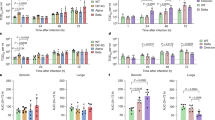
To examine whether the Omicron variant is also attenuated in vivo, we first compared virus propagation of Omicron with SARS-CoV-2 WT and the Delta variant in the K18-hACE2 transgenic mouse model. hACE2 transgenic mice (aged 6–8 weeks) were intranasally challenged with SARS-CoV-2 WT, Delta or Omicron, and their tissues were collected for virological assessment. Our results demonstrated that the infection of WT SARS-CoV-2 was very robust along the respiratory tract of the hACE2 transgenic mice as evidenced by the high RdRp gene copy numbers and the Delta variant propagated to a largely comparable level (Fig. 3a). By contrast, the viral RdRp gene copies in the nasal turbinate of mice challenged with Omicron were markedly reduced by 1,138-fold (P = 0.0324) and 1,821-fold (P = 0.001) compared with those that were challenged with WT and Delta, respectively, at 2 d post-infection (d.p.i.) (Fig. 3a), despite using the same inoculation titres for virus challenge among the aforementioned variants. At 4 d.p.i., the viral gene copies in the Omicron-infected mouse nasal turbinate were reduced by approximately tenfold compared with mice that were infected with the WT or Delta variant. Similarly, the RdRp gene copies were also significantly lower in the lungs of the mice infected with Omicron at both time points compared with their counterparts that were challenged with the WT or Delta variant (Fig. 3a).
Female and male K18-hACE2 transgenic mice (aged 6–8 weeks) were intranasally inoculated with 2 × 103 plaque-forming units (p.f.u.) of Alpha, Beta, Delta, Omicron or WT SARS-CoV-2. Nasal turbinate and lung samples of the infected mice were collected at 2 or 4 d.p.i. to determine the viral burden. n = 6 (WT and Delta) and n = 8 (Omicron). The body weight and survival of the infected mice were monitored for 14 days. n = 7 (Omicron), n = 15 (WT), n = 9 (Alpha and Delta) and n = 12 (Beta). a, Viral RNA-dependent RNA polymerase (RdRp) gene copies were quantified using probe-specific RT–qPCR. b, sgE expression was quantified using probe-specific RT–qPCR. c, Infectious viral titres were quantified using plaque assays in VeroE6-TMPRSS2 cells. d, The expression of the inflammatory cytokine genes Cxcl10 and Ifng was quantified using RT–qPCR. e, f, The body weight (e) and survival (f) of mice infected with SARS-CoV-2 WT, Alpha, Beta, Delta or Omicron. Data are mean ± s.d. from the indicated number of biological replicates. The colour key in e also applies to f. Statistical significance was determined using one-way ANOVA (a–d), two-tailed Student’s t-tests (e) and log-rank (Mantel–Cox) tests (f); where multiple comparisons were performed among different groups, P values were adjusted using Tukey’s multiple-comparison test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; NS, not significant. Data were obtained from two independent experiments. Each data point represents one biological replicate.
To understand whether the reduction in viral gene production was due to attenuated virus replication, we quantified the sgRNA of the SARS-CoV-2 E gene. Our results showed that the subgenomic E gene in the nasal turbinate of the Omicron-infected mice was significantly lower than that of the WT and Delta variant at both 2 d.p.i. and 4 d.p.i. (Fig. 3b). In particular, at 2 d.p.i., the sgRNA level was diminished by 442-fold (P = 0.0129) and 1,450-fold (P < 0.0001) compared with mice that were infected with the WT and Delta variant, respectively (Fig. 3b). Consistently, sgRNA synthesis was also lower in the lungs of Omicron-infected mice compared with mice infected with the WT or Delta variant, although to a lesser extent than in nasal turbinates (Fig. 3b). Consistent with the RdRp and subgenomic E gene results, the infectious virus titre in both nasal turbinate and lung samples of Omicron-infected mice was significantly lower compared with that of the WT-infected mice (Fig. 3c). Interestingly, the infectious virus titre in the nasal turbinate of Omicron-infected mice was 48-fold (P = 0.0476) lower than that of the Delta variant, suggesting less infectious virus shedding from the Omicron-infected nasopharynx compared with that from the Delta-infected nasopharynx at 2 d.p.i. (Fig. 3c). Consistently, viral gene copies in the nasal wash that were collected from the Omicron-infected transgenic mice were also reduced compared with samples that were collected from the other two groups (Extended Data Fig. 3). As the Omicron variant also carries the N501Y mutation, which enables the infection of WT mice and rats as we and others previously reported20,21, we also characterized the replication of the N501Y-carrying Alpha and Omicron variant in WT C57B6 mice side by side. Consistent with the findings in hACE2 transgenic mice, the replication of the Omicron variant in nasal turbinates and lungs of WT C57B6 mice was significantly attenuated compared with that of the Alpha variant (Extended Data Fig. 4). In parallel with the virological assessments, we quantified the gene expression of Cxcl10 (encoding IP-10) and Ifng (encoding IFNγ), which are hallmark proinflammatory cytokines induced by SARS-CoV-2 infection22,23. We found that the transcription levels of Cxcl10 and Ifng in both the nasal turbinates and lungs of Omicron-infected mice were significantly downregulated compared with mice that were challenged with the WT and Delta variant, which represented an alleviated pro-inflammatory response during the infection by the Omicron variant (Fig. 3d).
Next, we investigated the infection outcome of infection with Omicron compared with SARS-CoV-2 WT, Alpha, Beta and Delta in K18-hACE2 mice. Our results showed that, compared with the WT and the previously emerged variants, the body-weight loss of mice infected with Omicron was significantly milder, with the onset time at a later stage during the course of infection (Fig. 3e). Importantly, we took advantage of the K18-hACE2 infection model and compared animal survival after infection with SARS-CoV-2 WT and variants. Under the same inoculum conditions, mouse survival was lowest for Alpha (0%), followed by WT (20%), Beta (33%), Delta (44%) and was highest for Omicron (57%) (Fig. 3f). Statistically, mouse survival for Omicron was significantly higher than Alpha (P = 0.0001) and WT (P = 0.0377), but did not reach significance for Beta and Delta. Nevertheless, the trend of mouse survival was consistent with the results of body-weight loss, which demonstrated a propensity for better survival in the order of Alpha, WT, Beta, Delta and Omicron (Fig. 3f).
To evaluate the pathogenicity of Omicron infection in the respiratory tract, we performed histopathological analysis of the lung tissue of infected hACE2 transgenic mice. In corroboration with findings in the virological assessment, multifocal expression of the viral nucleocapsid (N) protein was frequently detected in both WT-infected and Delta-infected mice, but not in those that were challenged with the Omicron variant at 2 d.p.i. (Fig. 4a and Extended Data Fig. 5). At 4 d.p.i., abundant N protein continued to accumulate in the alveoli of mice infected with the WT and Delta variant. However, only a small amount of N protein expression was sparsely detected in the lungs of mice infected with the Omicron variant (Fig. 4a and Extended Data Fig. 5). Histological examination of both WT-infected and Delta-infected mouse lungs revealed prominent pathological changes in the alveoli, including the collapse of the alveoli wall, proteinaceous exudation in the alveoli cavity and epithelial damage in the small bronchioles, while interstitial congestion was commonly observed, most likely contributed by infiltrations of inflammatory cells and mononuclear lymphocytes (Fig. 4b and Extended Data Fig. 6). By contrast, histopathological changes in the lungs of mice infected with the Omicron variant were not evident at 2 d.p.i.. At 4 d.p.i., localized inflammatory infiltrations leading to thickening of the alveoli septa could be scarcely detected (Fig. 4b and Extended Data Fig. 6). Semi-quantitative histological scoring also indicated attenuated histopathological changes in the lungs of Omicron-infected mice compared with those infected with WT or Delta at both time points examined (Extended Data Fig. 7). Together, our results indicate that the Omicron variant replicates inefficiently in lung cells due to reduced spike cleavage and TMPRSS2 usage. The reduced replication ability of the Omicron variant results in attenuated lung pathology, milder body-weight loss and improved animal survival in vivo.
Female and male K18-hACE2 transgenic mice (aged 6–8 weeks) were intranasally inoculated with 2 × 103 p.f.u. of Delta, Omicron or WT SARS-CoV-2. The lungs of the infected mice were collected on 2 or 4 d.p.i. for histological analysis. a, Representative images of immunohistochemistry staining for the detection of nucleocapsid protein (brown, indicated by black arrows) of SARS-CoV-2 in the lungs of the infected mice. n = 3 (WT and Delta) and n = 4 (Omicron).b, Representative images of haematoxylin and eosin (H&E) staining for the detection of pathological tissue damage in the lungs of the infected mice. The images in a and b are representative images from three to four mice. n = 3 (WT and Delta) and n = 4 (Omicron). Dashed circles, mononuclear cell infiltration; dashed boxes, alveolar collapse; open arrowheads, interstitial congestion; closed arrowheads, perivascular infiltration; asterisk, proteinaceous exudation; cross, epithelium damage. Data were obtained from two independent experiments. Four to six sections were taken from each mouse for histology and immunochemistry analysis. For a and b, scale bars, 200 µm.
Discussion
The exact change of the Omicron variant that results in its severely impaired replication in Calu3 human lung epithelial cells and the respiratory tract of mice is currently unknown. Interestingly, our live-virus infection assays demonstrate that, although the replication of Omicron variant is substantially attenuated in Calu3 and Caco2 cells, it is less affected in VeroE6 cells. Our results are in agreement with a recent preprint suggesting that Omicron-pseudovirus entry is decreased in Calu3 and Caco2 cells, but not in Vero, HEK293T or A549-ACE2 cells16, as well as recent findings demonstrating the inefficient infection of human lung cells and organoids by Omicron24,25,26 . Mechanistically, we further demonstrate that the Omicron pseudovirus does not use TMPRSS2 for entry as efficiently as SARS-CoV-2 WT, Alpha, Beta or Delta pseudoviruses. At the same time, the inhibitory effect on the entry and replication of Omicron in VeroE6-TMPRSS2 cells by the TMPRSS2 inhibitor camostat is significantly weakened compared with the inhibitory effect of camostat on the entry and replication of the WT or the other variants. SARS-CoV-2 spike contains a PRRA insertion at the S1/S2 junction that creates a multibasic furin-like cleavage site. This motif is absent from the spike of SARS-CoV-1, SARS-CoV-related coronaviruses and the closely related bat coronavirus RaTG13 (ref. 27). Recent reports suggest that this acquired furin-like cleavage site facilitates the use of TMPRSS2 (refs. 28,29,30) and is critical for the highly efficient infection of human lung cells17,31,32. Our western blot analysis of virus-infected VeroE6-TMPRSS2 cells reveals decreased S1/S2 cleavage of Omicron spike compared with that of the SARS-CoV-2 WT and Delta variant. Prior S1/S2 cleavage facilitates the exposure of the S2′ cleavage site upon receptor binding that is required for TMPRSS2-mediated cleavage at S2′ and cell fusion33. Together, these results suggest that the attenuated replication of the Omicron variant in lung cells is due to a reduced efficiency of spike cleavage by host proteases owing to changes in the spike protein. Importantly, Omicron spike contains five substitutions at or near to the furin-like cleavage site, including T547K, D614G, H655Y, N679K and P681H, as well as six substitutions in S2, including N764K, D796Y, N856K, Q954H, N969K and L981F. Four of these substitutions (T547K, N764K, N856K and N969K) introduce electrostatic contacts formed between core S2 subunit helices and the S1 subunit34. Thus, the enhanced interactions between the S1 and S2 subunits in Omicron spike, along with substitutions at or near the furin-like cleavage site, may result in the altered S1/S2 cleavage. The exact role of these substitutions in S1/S2 cleavage should be further investigated.
A large body of timely reports has revealed that the Omicron variant can robustly escape neutralization antibodies, enabling it to infect fully vaccinated or even booster-vaccinated individuals2,3,4,5,6,7,8,9,10,13,14. Our results in this study suggest that the replication fitness of the Omicron variant is severely compromised potentially as a trade-off of escaping from neutralization antibodies. The SARS-CoV-2 variants that emerged before Omicron were known to have acquired mutations in spike that modulated their propagation in the human respiratory tract and the efficiency of virus transmission35,36,37. By comparing the pathogenicity of SARS-CoV-2 WT and the Alpha, Beta, Delta and Omicron variants side by side in the K18-hACE2 transgenic mouse model, we observed an attenuating trend for the emerging variants, with the Omicron variant being the mildest. The lower pathogenicity in mouse lungs infected with the Omicron variant hints that patients infected with the Omicron variant might develop milder respiratory symptoms compared with those after infection with previous variants.
Our study has limitations, including the use of the K18-hACE2 mouse model for pathogenesis studies instead of the primate models that are more similar to humans. However, the K18-hACE2 mouse model is a well-established model for SARS-CoV-2 study that enables the measurement of survival. Using this lethal model, we were able to compare the survival of mice after infection by SARS-CoV-2 WT, Omicron and the other variants side by side in a quantitative manner, which is not possible by using other non-human primate, hamster or ferret models.
Our study has a number of implications. First, we show that the Omicron variant is impaired in virus replication in human lung cells as well as in the lungs of infected animals, suggesting that Omicron infections in the human population may potentially result in milder lung symptoms compared with those after infection with previous variants. However, the Calu3 cells used in this study are carcinoma cells derived from the human lung and may not be accurate representations of the human lungs. Similarly, the K18-hACE2 mice may not fully resemble the pathogenesis of Omicron infection in humans. Moreover, data from our mouse models suggest that the difference in virus replication and pathogenicity between Omicron and Delta is smaller than the difference between Omicron and WT SARS-CoV-2. Thus, the pathogenicity of Omicron in humans should be further evaluated with human clinical data. Second, we show that the impaired replication of the Omicron variant results from its decreased spike cleavage and its ability to use TMPRSS2. In this regard, our results suggest that therapeutics targeting TMPRSS2 may be less effective against the Omicron variant. Overall, our study reveals key features of the Omicron variant that provide critical information for the prevention, control and treatment of the current COVID-19 pandemic.
Methods
Viruses and safety
WT SARS-CoV-2 HKU-001a (GenBank: MT230904), B.1.1.7/Alpha (GenBank: OM212469), B.1.351/Beta (GenBank: OM212470), B.1.617.2/Delta (GenBank: OM212471), B.1.1.529/Omicron (GenBank: OM212472) and B.1.1.529/Omicron(R346K) (GenBank: OM212473) were isolated from patients with laboratory confirmed COVID-19 in Hong Kong4,38. All variants of SARS-CoV-2 were cultured and titrated by plaque assays using VeroE6-TMPRSS2 cells. After obtaining the virus culture, the sequences of all variants used in this study were confirmed with nanopore sequencing. In vivo and in vitro experiments with infectious SARS-CoV-2 were performed according to the approved standard operating procedures of the Biosafety Level 3 facility at Department of Microbiology, HKU.
Cell cultures
Caco2, HEK293T and VeroE6 were obtained from ATCC and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) according to the supplier’s instructions. Calu3 cells were obtained from ATCC and maintained in DMEM/F12 (Gibco). VeroE6-TMPRSS2 cells were obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank and cultured in DMEM. All of the cell lines used are routinely tested for mycoplasma and are maintained mycoplasma-free.
In vivo virus challenge in mice
The use of animals was approved by the Committee on the Use of Live Animals in Teaching and Research of The University of Hong Kong under CULATR 5440-20 and 5370-20. Heterogenous K18-hACE2 C57BL/6J mice (2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from The Jackson Laboratory. The mice were kept in cages with individual ventilation under 65% humidity and an ambient temperature of 21–23 °C and a 12 h–12 h day–night cycle for housing and husbandry. Group sizes were chosen based on statistical power analysis and our prior experience in examining viral titres in SARS-CoV-2-infected K18-hACE2 transgenic and C57B6 mice. Experiments were repeated to give a sample size of 6 or above. Gender- and age-matched mice were randomized into different experimental groups. For virus challenge in mice, female C57BL/6J mice (aged 6–8 weeks) or female and male K18-hACE2 transgenic mice (aged 6–8 weeks) were anaesthetized with ketamine and xylazine, followed by intranasal inoculation with 20 µl per mouse of Alpha, Beta, Delta or Omicron variants or WT SARS-CoV-2 at 2 × 103 p.f.u. per mouse (for K18-hACE2 transgenic mice) or 1 × 105 p.f.u. per mouse (for C57BL/6J mice) as we previously described20,39. Mice were euthanized at 2 and 4 d.p.i. for collecting nasal turbinate and lung tissues for virological assessment, proinflammatory cytokine quantification or histological examination. Nasal wash was collected by flushing with 200 µl PBS on a daily basis for 4 days after virus challenge for viral-load detection. The survival and body weight of the infected animals were monitored for 14 days or until death of the animal.
Histology and immunohistochemistry staining
Animal tissues were collected and fixed with 10% neutral-buffered formalin. Nasal turbinates were decalcified with 10% formic acid for 7 days before being processed with the TP1020 Leica semi-enclosed benchtop tissue processor. IHC was performed with the DAB (3,3′-diaminobenzidine) substrate kit (Vector Laboratories) as we previously described39. To detect the viral antigen, in-house mouse monoclonal biotinylated anti-SARS-CoV-2 nucleocapsid protein antibodies (1:4,000) were used, followed by colour development using the DAB substrate kit. The nuclei were detected with haematoxylin before the tissue sections were mounted using the VectaMount permanent mounting medium (Vector Laboratories). For H&E staining, tissue sections were stained with Gill’s haematoxylin and eosin-Y. Images were acquired using the Olympus BX53 light microscope. Three to four mice were sampled in each group (as specified in the figure legends) and four to six sections from each animal were used for histology analysis. To obtain the semiquantitative histology scoring, tissue sections were graded in a blinded manner according to the pathological changes in bronchioles, alveoli and blood vessels by an experienced pathologist as previously reported by us and others40,41.
Infectious virus titration by plaque assays
Organs collected from infected mice were homogenized in DMEM using the Tissue Lyzer II (Qiagen) system and the clarified supernatants were tenfold serially diluted and inoculated to monolayered VeroE6-TMPSS2 cells for 2 h at 37 °C. After inoculation, the cells were washed with PBS three times, and covered with 1% low-melting agarose DMEM containing 1% FBS. The cells were fixed by 4% paraformaldehyde after 72 h incubation. The fixed samples were stained with 0.5% crystal violet in 25% ethanol/distilled water for plaque visualization.
Infectious virus titration by TCID50 assays
Supernatants from cells infected with the Alpha, Beta, Delta, Omicron variant, or WT SARS-CoV-2 were collected and tenfold serial diluted before inoculation into VeroE6-TMPRSS2 cells with four replicates per sample. Cytopathic effect was observed at 72 h.p.i. for the quantification of the TCID50.
Cell viability assays
VeroE6-TMPRSS2 cells were infected with the Alpha, Beta, Delta or Omicron variant, or WT SARS-CoV-2 at 0.1 m.o.i. Cell viability was quantified using the CellTiter-Glo luminescent cell viability assay kit (Promega) according to the manufacturer’s manual with the EnSight Multimode Microplate Reader (Perkin Elmer) at the designated time points.
Production of SARS-CoV-2-spike pseudoviruses and pseudovirus entry assays
All variants of SARS-CoV-2-spike pseudoviruses were packaged as described previously31,42. In brief, HEK293T cells were transfected with different spikes using Lipofectamine 3000 (Thermo Fisher Scientific). At 24 h after transfection, the cells were transduced with VSV-deltaG-firefly pseudotyped with VSV-G. At 2 h after transduction, the cells were washed three times with PBS and cultured in fresh medium with anti-VSV-G (8G5F11) antibodies (EB0010, Kerafast) (1:1,000). The pseudoviruses were then collected 16 h after transduction and titrated with TCID50. For pseudovirus entry assays, the target cells were inoculated with pseudoviruses for 2 h and cultured in 1% FBS medium for 24 h, before being washed and lysed for detection of the luciferase signal using a luciferase assay system (E1501, Promega).
RNA extraction and RT–qPCR
Viral RNA from infected cells was extracted using QIAsymphony RNA Kit (931636, Qiagen). Viral RNA from mouse lung and nasal turbinate samples was extracted using the RNeasy Mini kit (74106, Qiagen). SARS-CoV-2 gene copies targeting the RNA-dependent RNA polymerase (RdRp) were quantified using the QuantiNova Probe RT-PCR Kit (208354, Qiagen). Viral subgenomic RNA was detected using primers targeting the E gene. The expression of Cxcl10 and Ifng were detected using RT–qPCR using the QuantiNova SYBR Green RT-PCR kit. The primer and probe sequences are available on request.
Protease-inhibitor treatment assay
The serine protease inhibitor, camostat, was purchased from MedChemExpress. VeroE6-TMPRSS2 cells were treated with DMSO or camostat at concentrations of 1, 25, and 50 µM for 2 h before virus infection or pseudovirus transduction. At 24 h.p.i., the cell lysates were collected for RT–qPCR quantification of virus replication or lysed for detection of the luciferase signal.
Western blot analysis of spike cleavage from virus-infected VeroE6-TMPRSS2 cells
VeroE6-TMPRSS2 cells were infected by SARS-CoV-2 WT, Delta or Omicron at 0.01 m.o.i. Cell lysates were collected at 48 h.p.i. for western blot analysis. Specific primary antibodies were incubated with the blocked membranes at 4 °C overnight, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (Thermo Fisher Scientific) for 1 h at room temperature. The signal was developed using the SuperSignal West Pico PLUS Chemiluminescent Substrate (34580, Thermo Fisher Scientific) and detected using the Alliance Imager apparatus (Uvitec). Full-length spike and S2 was detected using rabbit anti-SARS-CoV-2 spike S2 antibodies (40590-T62, Sino Biological) (1:5,000). Nucleocapid (N) was detected using a custom rabbit anti-SARS-CoV-2 N immune serum (1:10,000) and β-actin was detected using anti-β-actin antibodies (AC-74, A5316, Sigma-Aldrich) (1:5,000).
Statistical analysis
Statistical comparisons between two experimental groups were performed using unpaired two-tailed Student’s t-tests. Comparisons among three or more experimental groups were performed using one-way or two-way ANOVA with Tukey’s multiple-comparison test. AUC values were calculated and analysed using one-way ANOVA. The survival of animals was compared using the log-rank (Mantel–Cox) test. Differences were considered to be statistically significant when P < 0.05. Data analysis was performed using GraphPad Prism v.8.0.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The complete sequence of SARS-CoV-2 HKU-001a (GenBank: MT230904), B.1.1.7/Alpha (GenBank: OM212469), B.1.351/Beta (GenBank: OM212470), B.1.617.2/Delta (GenBank: OM212471), B.1.1.529/Omicron (GenBank: OM212472), and B.1.1.529/Omicron(R346K) (GenBank: OM212473) are available on GenBank Source data are provided with this paper.
References
Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern (WHO, 2021); https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
Cele, S., Jackson, L., Khan, K., Khoury, D. & Sigal, A. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature https://doi.org/10.1038/s41586-021-04387-1 (2021).
Wang, Y. et al. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg. Microbes Infect. 11, 1–5 (2022).
Lu, L. et al. Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciab1041 (2021).
Dejnirattisai, W. et al. Reduced neutralisation of SARS-CoV-2 Omicron B.1.1.529 variant by post-immunisation serum. Lancet 399, 234–236 (2022).
Edara, V. et al. mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 Omicron variant. Preprint at https://doi.org/10.1101/2021.12.20.473557 (2021).
Ai, J. et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 11, 337–343 (2021).
Cao, Y. R. et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature https://doi.org/10.1038/s41586-021-04385-3 (2021).
Liu, L. et al. Striking antibody evasion manifested by the Omicron Variant of SARS-CoV-2. Nature https://doi.org/10.1038/s41586-021-04388-0 (2021).
VanBlargan, L. A., Errico, J. M., Halfmann, P., Zost, S. J. & Diamond, M. S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies. Preprint at https://doi.org/10.1101/2021.12.15.472828 (2021).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Chan, J. F. et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395, 514–523 (2020).
Espenhain, L. et al. Epidemiological characterisation of the first 785 SARS-CoV-2 Omicron variant cases in Denmark, December 2021. Euro Surveill. 26, 2101146 (2021).
Brandal, L. T. et al. Outbreak caused by the SARS-CoV-2 Omicron variant in Norway, November to December 2021. Euro Surveill. 26, 2101147 (2021).
Team, C. C.-R. SARS-CoV-2 B.1.1.529 (Omicron) variant - United States, December 1-8, 2021. MMWR Morb. Mortal Wkly Rep. 70, 1731–1734 (2021).
Hoffmann, M. et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell 185, 447–456.e411 (2022).
Hoffmann, M., Kleine-Weber, H. & Pöhlmann, S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78, 779–784 (2020).
Koch, J. et al. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J. 40, e107821 (2021).
Beumer, J. et al. A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat. Commun. 12, 5498 (2021).
Shuai, H. et al. Emerging SARS-CoV-2 variants expand species tropism to murines. eBioMedicine 73, 103643 (2021).
Gu, H. et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369, 1603–1607 (2020).
Laing, A. G. et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 26, 1623–1635 (2020).
Diorio, C. et al. Multisystem inflammatory syndrome in children and COVID-19 are distinct presentations of SARS-CoV-2. J. Clin. Invest. 130, 5967–5975 (2020).
Zeng, C. et al. Neutralization and Stability of SARS-CoV-2 Omicron Variant. Preprint at https://doi.org/10.1101/2021.12.16.472934 (2021).
Meng, B. et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature https://doi.org/10.1038/s41586-022-04474-x(2022).
Zhao, H. et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with delta variant in TMPRSS2-expressed cells. Emerg. Microbes Infect. 11, 277–283 (2022).
Coutard, B. et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 176, 104742 (2020).
Mykytyn, A. Z. et al. SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. eLife 10, e64508 (2021).
Lamers, M. M. et al. Human airway cells prevent SARS-CoV-2 multibasic cleavage site cell culture adaptation. eLife 10, e66815 (2021).
Laporte, M. et al. The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. PLoS Pathog. 17, e1009500 (2021).
Chu, H. et al. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat. Commun. 12, 134 (2021).
Peacock, T. P. et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 6, 899–909 (2021).
Park, J. E. et al. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl Acad. Sci. USA 113, 12262–12267 (2016).
McCallum, M. et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Preprint at https://doi.org/10.1101/2021.12.28.474380 (2021).
Mlcochova, P. et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599, 114–119 (2021).
Liu, Y. et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Preprint at https://doi.org/10.1101/2021.08.12.456173 (2021).
Saito, A. et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 602, 300–306 (2022).
Chu, H. et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe 1, e14–e23 (2020).
Chu, H. et al. Targeting highly pathogenic coronavirus-induced apoptosis reduces viral pathogenesis and disease severity. Sci. Adv. 7, eabf8577 (2021).
Yuan, S. et al. Clofazimine broadly inhibits coronaviruses including SARS-CoV-2. Nature 593, 418–423 (2021).
Zheng, J. et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589, 603–607 (2021).
Havranek, K. E. et al. SARS-CoV-2 spike alterations enhance pseudoparticle titers and replication-competent VSV-SARS-CoV-2 virus. Viruses 12, 1465 (2020).
Acknowledgements
This work was supported in part by funding from the Health and Medical Research Fund (CID-HKU1-5, COVID1903010, projects 7 and 14, and 20190652), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region; the General Research Fund (17118621) and the Theme-Based Research Scheme (T11-709/21-N) of Research Grants Council, The Government of the Hong Kong Special Administrative Region; Health@InnoHK, Innovation and Technology Commission, the Government of the Hong Kong Special Administrative Region; National Natural Science Foundation of China Excellent Young Scientists Fund (Hong Kong and Macau) (32122001); National Program on Key Research Project of China (grant no. 2020YFA0707500 and 2020YFA0707504); the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for Department of Health of the Hong Kong Special Administrative Region Government, Sanming Project of Medicine in Shenzhen, China (no. SZSM201911014); the High Level-Hospital Program, Health Commission of Guangdong Province, China; the University of Hong Kong Li Ka Shing Faculty of Medicine Enhanced New Staff Start-up Fund; the University of Hong Kong Outstanding Young Researcher Award; the University of Hong Kong Li Ka Shing Faculty of Medicine Research Output Prize; the Major Science and Technology Program of Hainan Province (ZDKJ202003); the research project of Hainan Academician Innovation Platform (YSPTZX202004); the Hainan Talent Development Project (SRC200003); the Emergency Key Program of Guangzhou Laboratory (EKPG22-01); and the donations of the Shaw Foundation Hong Kong, R. Yu and C. Yu, M. T. M. M. Yin, M. S.-K. Tong, the Providence Foundation Limited (in memory of the late L. H. Minh), Lee Wan Keung Charity Foundation Limited, H. Ming, H. Hoy and Chow Sin Lan Charity Fund Limited, Hong Kong Sanatorium & Hospital, the Chan Yin Chuen Memorial Charitable Foundation, M. M.-W. Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, the Jessie & George Ho Charitable Foundation, Perfect Shape Medical Limited, K. C. Tong, Foo Oi Foundation Limited, T. K. M. Laurence, B. H.-C. Lee, P. C. So and the Lo Ying Shek Chi Wai Foundation. The funding sources had no role in the study design, data collection, analysis, interpretation or writing of the report.
Author information
Authors and Affiliations
Contributions
Conceptualization: H.S., J.F.-W.C., B.H., Y.C., K.-Y.Y. and H.C. Methodology: H.S., B.H., Y.C., T.T.-T.Y. and H.C. Investigation: H.S., B.H., Y.C., T.T.-T.Y., F.Y., X.H., C.Y., J.-C.H., H.L., J.S., Y.L., T.Z., J. Zhang, Y.H., Y.W., L.L., J.-P.C., A.J.Z., J. Zhou, S.Y., M.A.B., B.-Z.Z., J.-D.H. and K.K.-W.T. Visualization: H.S., B.H., Y.C. and H.C. Funding acquisition: J.F.-W.C., F.Y., K.-Y.Y. and H.C. Project administration: J.F.-W.C., K.-Y.Y. and H.C. Supervision: J.F.-W.C., K.-Y.Y. and H.C. Writing—original draft: H.S., J.F.-W.C., B.H., Y.C., K.-Y.Y. and H.C. Writing—review and editing: H.S., J.F.-W.C., B.H., Y.C., K.-Y.Y. and H.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
thanks Emmie de Wit and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Virus replication of Omicron, WT SARS-CoV-2, and other VOCs in Caco2 and VeroE6 cells.
a Caco2 and b VeroE6 cells were challenged by SARS-CoV-2 WT, Alpha, Beta, Delta, Omicron, or Omicron(R346K) at 0.1 m.o.i. Cell lysates were harvested at the designated time points for the quantification of subgenomic RNA of the envelope (sgE) gene. Robustness of sgE production was quantified with the AUC analysis (n = 7). Data represents mean ± SD from the indicated number of biological repeats. Statistical significances were determined with one-way ANOVA with Tukey’s multiple comparison tests. Each data point represents one biological repeat. * represented p < 0.05 and ** represented p < 0.01. *** represented p < 0.001, **** represented p < 0.0001. ns, not statistically significant.
Extended Data Fig. 2 Western blot analysis of SARS-CoV-2 spike from virus infected VeroE6-TMPRSS2 cells.
VeroE6-TMPRSS2 cells were infected by SARS-CoV-2 WT, Delta, or Omicron at 0.01 m.o.i. Cell lysates were harvested at 48 h post infection for detection of SARS-CoV-2 spike cleavage using an anti-spike S2 antibody. Representative images of SARS-CoV-2 spike and nucleocapsid Western blots were shown with β-actin added as a sample processing control. Spike, nucleocapsid, and β-actin was run on three different gels and detected on three different membranes. The experiment was repeated three times independently with similar results.
Extended Data Fig. 3 Viral burden in nasal wash of K18-hACE2 transgenic mice infected with SARS-CoV-2 WT, Delta, or Omicron.
Male and female 6- to 8-week old K18-hACE2 mice were infected with 2 × 103 pfu SARS-CoV-2 WT, Delta, or Omicron (n = 4, 5 and 7 for WT, Delta and Omicron, respectively). Nasal wash was collected on a daily basis for four days for determining RdRp gene copies with RT-qPCR. Statistical significances were determined with one-way ANOVA. Multiple comparisons among different groups were adjusted with Tukey’s multiple comparison tests. Data were obtained from two independent experiments. Each data point represents one biological repeat. * represented p < 0.05. ns, not statistically significant.
Extended Data Fig. 4 Omicron infection in C57B6 WT mice.
6-to-8-week-old female C57B6 WT mice were intranasally inoculated with 1 × 105 PFU of Alpha or Omicron. Nasal turbinates and lungs of the infected mice were collected on 2 or 4 d.p.i. for viral burden determination (n = 8). a Viral RNA-dependent RNA polymerase (RdRp) gene copies were quantified with probe-specific RT-qPCR. b Subgenomic envelope (E) gene were quantified with probe-specific RT-qPCR. c Infectious viral titres were quantified with plaque assay in VeroE6-TMPRSS2 cells. Data represents mean ± SD from the indicated number of biological repeats. Statistical differences were determined with two-tailed Student’s t-test in (a–c). Data were obtained from two independent experiments. Each data point represents one biological repeat. * represented P < 0.05; ** represented P < 0.01; *** represented P < 0.001; **** represented P < 0.0001. ns, not statistically significant.
Extended Data Fig. 5 Immunohistochemistry staining of lung tissue of K18-hACE2 transgenic mice infected with SARS-CoV-2 WT, Delta, or Omicron.
6-to-8-week-old female and male K18-hACE2 transgenic mice were intranasally inoculated with 2 × 103 PFU SARS-CoV-2 WT, Delta, or Omicron. Lung of the infected mice were collected on 2 or 4 d.p.i. for histological analysis. Representative images of immunohistochemistry staining for the detection of nucleocapsid protein (brown, pointed by black arrows) of SARS-CoV-2 in lung of the infected mice (n = 3 for WT and Delta; n = 4 for Omicron). Images are the source images with lower magnification for Fig. 4a. Dashed rectangles indicate the region enlarged in Fig. 4a. Four to six sections were taken from each mouse for immunochemistry analysis. Scale bar represents 500 µm.
Extended Data Fig. 6 H&E images of lung tissue of K18-hACE2 transgenic mice infected with SARS-CoV-2 WT, Delta, or Omicron.
6-to-8-week-old female and male K18-hACE2 transgenic mice were intranasally inoculated with 2 × 103 PFU SARS-CoV-2 WT, Delta, or Omicron. Lung of the infected mice were collected at 2 or 4 d.p.i. for histological analysis. Representative images of H&E staining for the detection of pathological tissue damage in the nasal turbinate and lung of the infected mice (n = 3 for WT and Delta; n = 4 for Omicron). Images are the source images with lower magnification for Fig. 4b. Dashed rectangles indicate the region enlarged in Fig. 4b. Four to six sections were taken from each mouse for histology analysis. Scale bar represents 500 µm.
Extended Data Fig. 7 Histological scoring of lung tissue of K18-hACE2 transgenic mice infected with SARS-CoV-2 WT, Delta, or Omicron.
Semiquantitative histology scores were given to each H&E image of the lung tissue by grading the severity of damage in bronchioles, alveoli, and blood vessels and accumulating the total scores. For bronchioles: 0 = normal structure; 1 = mild peribronchiolar infiltration; 2 = peribronchiolar infiltration plus epithelial cell death; and 3 = score 2 plus intrabronchiolar wall infiltration and epithelium desquamation. For alveoli: 0 = normal structure; 1 = alveolar wall thickening and congestion; 2 = focal alveolar space infiltration or exudation; and 3 = diffuse alveolar space infiltration or exudation or haemorrhage. For blood vessels: 0 = normal structure; 1 = mild perivascular oedema or infiltration; 2 = vessel wall infiltration; and 3 = severe endothelium infiltration. Data are mean ± SD (n = 3 for WT and Delta; n = 4 for Omicron). Statistical differences were determined with one-way ANOVA. Multiple comparisons among different groups were adjusted with Tukey’s multiple comparison tests. Images were obtained from two independent in vivo experiments. Each data point represents one biological repeat. * represented P < 0.05; ** represented P < 0.01; *** represented P < 0.001; **** represented P < 0.0001. ns, not statistically significant.
Supplementary information
Rights and permissions
About this article
Cite this article
Shuai, H., Chan, J.FW., Hu, B. et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603, 693–699 (2022). https://doi.org/10.1038/s41586-022-04442-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04442-5