Abstract
Aim This systematic review sought to assess and scrutinise the validity and practicality of published and preprint reports of prediction models for the diagnosis of coronavirus disease 2019 (COVID-19) in patients with suspected infection, for prognosis of patients with COVID-19, and for identifying individuals in the general population at increased risk of infection with COVID-19 or being hospitalised with the illness.
Data sources A systematic, online search was conducted in PubMed and Embase. In order to do so, the authors used Ovid as the host platform for these two databases and also investigated bioRxiv, medRxiv and arXiv as repositories for the preprints of studies. A public living systematic review list of COVID-19-related studies was used as the baseline searching platform (Institute of Social and Preventive Medicine's repository for living evidence on COVID-19).
Study selection Studies which developed or validated a multivariable prediction model related to COVID-19 patients' data (individual level data) were included. The authors did not put any restrictions on the models included in their study regarding the model setting, prediction horizon or outcomes.
Data extraction and synthesis Checklists of critical appraisal and data extraction for systematic reviews of prediction modelling studies (CHARMS) and prediction model risk of bias assessment tool (PROBAST) were used to guide developing of a standardised data extraction form. Each model's predictive performance was extracted by using any summaries of discrimination and calibration. All these steps were done according to the aspects of the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) and preferred reporting items for systematic reviews and meta-analyses (PRISMA).
Results One hundred and forty-five prediction models (107 studies) were selected for data extraction and critical appraisal. The most common predictors of diagnosis and prognosis of COVID-19 were age, body temperature, lymphocyte count and lung imaging characteristics. Influenza-like symptoms and neutrophil count were regularly predictive in diagnostic models, while comorbidities, sex, C-reactive protein and creatinine were common prognostic items. C-indices (a measure of discrimination for models) ranged from 0.73 to 0.81 in prediction models for the general population, from 0.65 to more than 0.99 in diagnostic models, and from 0.68 to 0.99 in the prognostic models. All the included studies were reported to have high risks of bias.
Conclusions Overall, this study did not recommend applying any of the predictive models in clinical practice yet. High risk of bias, reporting problems and (probably) optimistic reported performances are all among the reasons for the previous conclusion. Prompt actions regarding accurate data sharing and international collaborations are required to achieve more rigorous prediction models for COVID-19.
Similar content being viewed by others
A commentary on
Wynants L, Van Calster B, Collins G S et al.
Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. BMJ 2020; 369: m1328.

GRADE rating
Commentary
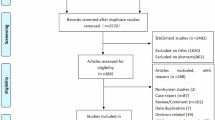
The paper aimed to review and critically assess the reliability and performance of current prediction models for COVID-19 while following the preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement. The authors succeeded in representing a coherent, up-to-date and systematically written paper according to PRISMA. Key findings of this study are represented as a concept map in Figure 1.
Studies from PubMed or Embase were considered as eligible if they developed or validated a multivariable model or estimation system, based on individual participant level data, to predict any COVID-19-related outcome. All epidemiological studies which sought to model disease transmission or case fatality rates, diagnostic test accuracy or predictor finding were excluded. The authors also contacted other studies' author teams for non-public results which fitted into their scope (two studies). Studies that were publicly available but not listed on the living systematic review list were further added to the data source (six studies). Since multiple rounds of searching and literature assessment have been performed, from the first round onwards, the gathered information was initially assessed using a text analysis tool developed by artificial intelligence to prioritise sensitivity.
The authors included 275 out of 14,217 titles to be reviewed by abstract and full text screening. From these, 145 prediction models (107 studies) were selected for data extraction and critical appraisal. These prediction models consisted of the following: 91 diagnostic models for verifying COVID-19 in suspected patients (both for the presence or severity of illness based on either medical imaging or diagnosing disease severity); 50 models for predicting the prognosis and trajectory of the disease in COVID-19 patients (which predicted the mortality risk, progression to severe disease, intensive care unit admission, ventilation, intubation or length of hospital stay); and four models for identifying individuals in the general population who are more susceptible to contracting COVID-19.
The mortality rate among the prognostic models predicting mortality risk in people with confirmed or suspected infection varied between 1% to 59%. The authors recognised a substantial sampling bias due to excluded participants who still had the disease at the end of each included study (that is, they had neither recovered nor died). Further, follow-up period was different among the studies (rarely reported), and there might also be local and temporal variations in the admittance criteria for hospitalised patients in each study or for getting diagnosed with COVID-19. The prevalence of COVID-19 ranged between 17% and 79% for the diagnostic studies, although the prevalence figures were deemed not to be representative of their target populations due to unclear data collection methodologies. No COVID-19 datasets were used in the models predicting risks of COVID-19 in general populations. However, some proxy outcomes (non-tuberculosis pneumonia, influenza, acute bronchitis or upper respiratory tract infections) were considered in these models. Age, sex, previous hospital admissions, comorbidity data and social determinants of health were considered as frequent predictors in these models. The highest C-index was reported as 0.81 in these four models.
In COVID-19 diagnostic models for patients with suspected infection, the reported C-index values varied between 0.65 and 0.99. Out of nine studies diagnosing severe disease in COVID-19-positive patients, one study was done on paediatric patients with reported perfect performance. Common predictors of severe COVID-19 were comorbidities, liver enzymes, C-reactive protein, imaging features and neutrophil count. In prognostic models for COVID-19-positive patients, the most common prognostic factors included comorbidities, age, sex, lymphocytic count, C-reactive protein, body temperature, creatinine and imaging features. Studies forecasting mortality rate reported C-indices between 0.68 and 0.98, while the models predicting progression to a severe or critical state (not mortality) reported C-indices between 0.73 and 0.99. According to the results of calibration and discrimination for these studies, the models were either evaluating the mortality risk as too high or too low, over-fitted or underestimated, or unclear with regards to the calibration methodology.
All the included studies were at high risk of bias when assessed using prediction model risk of bias assessment tool (PROBAST) protocol. This implied that the models' predictive performance was probably lower than what had been reported. Participation bias, unclear reporting, risk of bias predictor and analysis domains, and use of subjective or proxy outcomes were all suggested as causes for concern in these models. The authors also encouraged medical researchers to report more detailed information about the pre-processing steps of modelling studies (for example, cropping the medical images).
The majority of studies used data from China or public international data repositories. This was mentioned as a probable cause of over-fitted predictive models, which could lead to lower accuracy of performance when applied to new samples. Adjusting the data based on the local hospital admission protocols is another important aspect for future predictions to achieve more calibrated models. Limitations of the study were also noted as the authors could not consider the future amendments of unpublished studies (which could be improved through the review process).
This study reiterates the importance of critical thinking as a medical practitioner, whose job is not only to treat patients, but also includes providing patients with the most up-to-date and safest therapeutic options available.1 Data sharing while considering the privacy protection of patients is another issue presented as an urgent need. This could ultimately increase the generalisability and implementation of COVID-19 predictive models when applied to different populations and settings during the external validation process. Mathematical prediction models could be of utmost importance, given that accurate epidemiological estimations could lead to more effective management strategies during public health emergencies.2 Another major concern is the lack of robust prediction evidence in paediatrics. As is mentioned in this review, only one study had developed prediction models for use in paediatric patients. Given that children could majorly affect the spread of COVID-19, it is highly recommended to provide more evidence in this regard.3
Overall, the current results of this living systematic review are in line with the literature about the validity of predictive models regarding the detection and prognosis of COVID-19.4,5 While appreciating the scientific race of COVID-19 predictive papers in medicine, developing more rigorous models using the previously published studies should be of more concern.
References
Gupta M, Upshur R. Critical thinking in clinical medicine: what is it? J Eval Clin Pract 2012; 18: 938-944.
Zhong L, Mu L, Li J, Wang J, Yin Z, Liu D. Early prediction of the 2019 novel coronavirus outbreak in the mainland china based on simple mathematical model. IEEE Access 2020; DOI: 10.1109/ACCESS.2020.2979599.
Mallapaty S. How do children spread the coronavirus? The science still isn't clear. Nature 2020; 581: 127-128.
Agbehadji I E, Awuzie B O, Ngowi A B, Millham R C. Review of Big Data Analytics, Artificial Intelligence and Nature-Inspired Computing Models towards Accurate Detection of COVID-19 Pandemic Cases and Contact Tracing. Int J Environ Res Public Health 2020; DOI: 10.3390/ijerph17155330.
Jewell N P, Lewnard J A, Jewell B L. Predictive Mathematical Models of the COVID-19 Pandemic: Underlying Principles and Value of Projections. JAMA 2020; DOI: 10.1001/jama.2020.6585.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Shamsoddin, E. Can medical practitioners rely on prediction models for COVID-19? A systematic review. Evid Based Dent 21, 84–86 (2020). https://doi.org/10.1038/s41432-020-0115-5
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41432-020-0115-5