Abstract
The reported new confirmed cases of Coronavirus Disease 2019 (COVID-19) nowadays have diminished in their usefulness for assessing the pandemic situation. This study aimed to discover the correlation of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) variants recorded by Nextstrain clade and PANGO lineage and the number of new confirmed cases. Percent stacked area charts were utilized to display their development trends. 31 and 1452 variants were named according to Nextstrain clade and PANGO lineage, respectively. The branch step value maintained a stable increase by linear regression analysis. The changing trend in SARS-CoV-2 variants (PANGO lineage) correlated negatively with the number of new confirmed COVID-19 cases through Spearman rank correlation coefficient (17/06/2020-17/11/2021, ρ=-0.387, P < 0.01; 15/12/2021-11/01/2023, ρ=-0.458, P < 0.01). The proportion and composition of dominant virus variants had regional discrepancies, but some also fluctuated. The speed and quantity of SARS-CoV-2 variants objectively reflect the characteristics of the COVID-19 pandemic and viral dissemination in the population even without valuable data of reported new confirmed cases. The SARS-CoV-2 variation may be a better tool for epidemic monitoring and early-warning in the low epidemic state.
Similar content being viewed by others
Introduction
The coronavirus disease 2019 (COVID-19) pandemic began since December 20191. The control and prevention measures worldwide were lifted progressively, such as mass rapid testing, mask use and vaccination campaigns2. World Health Organization (WHO) determined that COVID-19 is an established and ongoing health issue that no longer constitutes a public health emergency of international concern (PHEIC) in May 20233. However, whether the pandemic has come to an end has provoked heated discussion among scientists and people, which has not yet reached an acceptable or quantitative definition4,5,6,7. The establishment of immunity by infection or vaccination, the reduction in daily new confirmed cases and the proportion of severe cases, the restoration of medical system function, and the decrease in infection risk, are all perceptible signs of the end of the pandemic. In terms of the epidemiology of infectious diseases, a reproduction number less than 1 is generally considered to indicate that an epidemic is controlled and nearing its end8,9. However, new COVID-19 cases are no longer evaluated or reported, so little evidence can be inferred from the current pandemic data, and it is difficult to obtain the exact value of the reproduction number.
Many studies predicted the pandemic trend of COVID-19 based on compartment model or machine learning models with timely COVID-19 data. The former need to remodify its differential equations frequently considering the epidemic spread dynamics, which increased the complexity of model application10,11; The latter, widely used in the healthcare and environment, is more accurate but require sufficient data12. Unfortunately, they were useless without timely COVID-19 data in the low epidemic state. However, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants are still monitored and recorded at the GISAID website. For example, the Delta variant of SARS-CoV-2, firstly discovered in India in October 2020, has the characteristics of stronger infectivity, higher viral load, and shorter incubation period, threatening nearly 200 countries and regions13. Omicron, firstly detected in South Africa in November 2021, spreads faster than Delta with low virulent and still occupies the ongoing dominant strain14. Moreover, there is no reported study to explore the relationship between the SARS-CoV-2 variants and COVID-19 pandemic trend. Nowadays SARS-CoV-2 variants are mainly named by the Nextstrain clade and the PANGO lineage15,16,17,18: the Nextstrain clade is an open-source project for viral genome alignment, mutation calling, clade assignment, quality checks and phylogenetic placement; the PANGO lineage is a dynamic nomenclature system for tracking the transmission and spread of SARS-CoV-2, including variants of concern. This paper analyzed the characteristics of the relationships between variant strains, recorded by the Nextstrain clade and the PANGO lineage, and epidemic data collected during the three years of the COVID-19 pandemic, which may determine the current situation by monitoring SARS-CoV-2 variants. This paper aimed to explore a feasible path to real-time monitoring and early warning in an objective and quantitative manner during the low epidemic stage.
Data and methods
COVID-19 pandemic data were derived from the GitHub website (https://github.com/CSSEGISandData/COVID-19/tree/master/csse_covid_19_data/csse_covid_19_daily_reports), which provides open-source and real-time epidemiological data for non-profit purpose. The prevalence ratio and number of SARS-CoV-2 variants, recorded by the two virus naming methods of the Nextstrain clade and the PANGO lineage, were obtained through the GISAID website (https://nextstrain.org/charon/getDataset? prefix=ncov%2Fgisaid%2Fglobal%2Fall-time&type=tip-frequencies), which promotes the rapid sharing of data from priority pathogens to help researchers understand how viruses evolve and spread during epidemics and pandemics. Since WHO announced that COVID-19 was no longer a PHEIC in May 2023 and many countries cancelled the control and prevention measures, the newly reported COVID-19 cases have decreased rapidly worldwide and lost the guidance value. However, this study extracted the meaningful data (January, 2020 to January, 2023) to analyze the correlation between SARS-CoV-2 variants and the new confirmed COVID-19 cases innovatively, in order to discover a potential method for monitoring and early-warning in the low epidemic state. The linear regression was adopted to investigate their relationship. Besides, Pearson correlation coefficient and Spearman rank correlation coefficient, as two classic methods for correlation analysis, were also considered. The scatter plot of new confirmed COVID-19 cases and PANGO lineage was drawn and followed by a K-S test to examine the normality. These indicated that Spearman rank correlation coefficient was more suitable and a significance test was conducted to prove its reliability. Here is the flowchart for the analysis process (Fig. 1).
Flowchart for the analysis process of this study. The data were derived from the GitHub and GISAID website from January, 2020 to January, 2023; the linear regression, Pearson correlation coefficient and Spearman rank correlation coefficient were adopted to analyze the correlation between SARS-CoV-2 variants and the new confirmed COVID-19 cases.
Results
SARS-CoV-2 variants were correlated with the status of the COVID-19 pandemic
As of January 25, 2023, a total of 31 and 1452 SARS-CoV-2 variants were named according to the Nextstrain clade and the PANGO lineage, respectively. Generally, a clade is designated if a variant shows a consistent growth of > 0.05 per day in frequency where it is circulating, in addition to reaching > 5% regional frequency. After the Omicron variant became widely prevalent and dominant, compared with the previous variant, it had an obvious suppressive effect on the epidemic transmission of other variants. It slowed the development of new branches and further extended its transmission time, which may be related to the fact that the reproduction number of the Omicron variant was similar to that of measles.
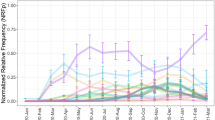
The trends of SARS-CoV-2 variants (Nextstrain clade) derived from the GISAID website were showed in Fig. 2 and further analyzed using the linear regression. The following regression equation was obtained (y denotes the branch step, which means the accumulation of SARS-CoV-2 variants at run time, and x denotes the date):
F-test was performed on the linear relationship between branch step (y) and time (x), where ŷ was the estimated value of y, and the F-value (4463.295, P < 0.001) demonstrated the significance of Eq. (1). The coefficient of determination (R2) is the ratio of the sum of squares of the regression to the sum of squares of the total deviation. R2 was calculated as 0.992, indicating that 99.2% of the variation of branch step can be explained by date. Moreover, the T-test is performed on the sample regression coefficient and constant terms (Table 1). The result indicated that branch step increases by 3.707 × 10-5 every day on average, demonstrating that the change of branch step depended on the date steadily and can be regarded as credible data source. Therefore, the branch step value presented a stable linear increase (Fig. 2), which indicated that the spread and number of viral mutations were stable and did not change regardless of the presence of different virus variants.
The number of new confirmed COVID-19 cases and the trend of SARS-CoV-2 variants (PANGO lineage), which come from the GitHub website and the GISAID website respectively, were further analyzed to illuminate the trend of the COVID-19 pandemic. A scatter plot was drawn using the data from January 1, 2020 to January 11, 2023 (Fig. 3A). The number of new confirmed cases decreased significantly along with the increase of the new SARS-CoV-2 variants (PANGO lineage), exhibiting an inversely linear trend. Pearson correlation coefficient, a common method for linear analysis, requires that the corresponding variables obey bivariate normal distribution. However, the number of new confirmed cases and the new PANGO lineage did not follow the normal distribution with K-S test. Instead, Spearman rank correlation coefficient (ρ), also known as rank correlation coefficient, was utilized to determine their relationship. Spearman rank correlation coefficient is a non-parametric statistical method and used widely to analyze the linear correlation of two variables.
Given the different transmissibility of SARS-CoV-2 variants, 3 intervals were divided to analyze their relationship through Spearman rank correlation coefficient (Table 2), including 1st interval (the whole time, 01/01/2020-11/01/2023), 2nd interval (Delta, 17/06/2020-17/11/2021), and 3rd interval (Omicron, 15/12/2021-11/01/2023). Reflected from the whole time (1st interval) (Fig. 3B and C), 2nd interval entered a relatively low epidemic state after the initial outbreak while 3rd interval exhibited a complete epidemic process (up-peak-down). There were of great significance for the relationship between new PANGO lineage and new confirmed cases in both 2nd and 3rd intervals, but there was no statistical significance of rank correlation coefficient in 1st interval (Table 2). In order of data fully utilization, the Pearson correlation coefficient under the ignorance of skewness is -0.157 (P < 0.05), showing a weak negative correlation between new PANGO lineage and new confirmed cases in 1st interval. The different transmissibility of variants affected the data of new confirmed cases, which might weaken their correlation in 1st interval to a certain extent.
The Delta strain was firstly reported in October 2020 and had become the dominant strain in many countries since July 2, 2021. The number of new confirmed cases in 2nd interval exhibited a relatively stable growth after the rapid increasing in 3 months from June 2020 (Fig. 3D). The new PANGO lineage negatively correlated to the number of new confirmed cases in 2nd interval according to the two regression trend lines. Besides, the divergence of the opposite trend was increasing. After one month, the Omicron variant gradually replaced the Delta and became another dominant strain, triggering the return of COVID-19 outbreak. The Omicron strain became the dominant strain, resulting in a larger-scale infection than Delta and then stepped into a low epidemic state in 3rd interval (Fig. 3E). There was an opposite correlation between the new PANGO lineage and the number of new confirmed cases, and the trend divergence was increasing. Many countries have abolished control and prevention measures since January 11, 2023 due to the decreasing number of new confirmed cases, corresponding with the data analysis of this study.
It was the different transmissibility of dominant strain that contributed to no significant correlation between the new PANGO lineage and the number of new confirmed cases in the whole time. However, the pandemic trend of new confirmed cases was obviously opposite with the new PANGO lineage during a period with specific dominant strain. The new PANGO lineage could provide certain reference value for pandemic monitoring and early-warning at the low epidemic state.
Negative relationship between SARS-CoV-2 variants and the number of confirmed COVID-19 cases. (A) The scatter plot of the relationship between the number of SARS-CoV-2 variants according to PANGO lineage and the number of new confirmed cases from January 1, 2020 to January 11, 2023, exhibiting an inversely linear trend; Weekly named variants according to the PANGO lineage (B) and the Nextstrain clade (C) with new confirmed cases in 1st interval (whole time, 01/01/2020-11/01/2023), indicating that 2nd interval (Delta, 17/06/2020-17/11/2021) entered a relatively low epidemic state after the initial outbreak while 3rd interval (Omicron, 15/12/2021-11/01/2023) a complete epidemic process (up-peak-down); Regression lines of new confirmed cases and new PANGO lineage in 2nd interval (D) and 3rd interval (E).
Worldwide epidemic progress and evolution of SARS-CoV-2 variants
Percent stacked area charts were utilized to show the prevalence proportion and distribution of SARS-CoV-2 variants worldwide at different times, by calculating the number of COVID-19 cases infected with different SARS-CoV-2 variants, according to the Nextstrain clade and PANGO lineage (Fig. 4). From September 2020 to September 2021, multiple SARS-CoV-2 variants were prevalent simultaneously worldwide, and the dominant variants changed frequently. After October 2021, each dominant variant (Nextstrain clade) was dominant for approximately 3 months and was then replaced. The dominant variants in different regions were not synchronous, and the proportions of some dominant variants fluctuated (Fig. 4A; Table 3). In addition, the trend of SARS-CoV-2 variants according to the PANGO lineage was similar to that according to the Nextstrain clade (Fig. 4B).
In January 2023, 22E (Omicron), 22 F (Omicron), 22D (Omicron), 22B (Omicron), and 21 L (Omicron) were the most globally prevalent and widely distributed strains, with proportions of 45.8%, 23.5%, 16.8%, 10.7% and 1.6%, respectively. The prevalent strains and their proportions were different in different areas, which showed the diversity of the pandemic situation (Table 3).
Discussion
According to the rules for the designation and naming of the PANGO lineage and the Nextstrain clade naming strategy for 2022, variants are given a name when they prevail in a large proportion of the infected population or for a long time with epidemiological relevance, especially mutations documented in the receptor-binding domain (RBD) of the viral spike protein that increases the viral spread rate or disease severity19,20,21. The named virus strain is called the “dominant virus strain”22. As viral mutations are random and instantaneous23, the speed and quantity of viral variants objectively reflect the characteristics of the SARS-CoV-2 epidemic and dissemination in the population and are the result of the comprehensive effects of various environmental factors, including the establishment of an immune barrier by infection or vaccination, social prevention and control measures, and cross-regional population mobility.
This study showed that fluctuations in the COVID-19 pandemic were correlated with the number and frequency of SARS-CoV-2 variants in the low epidemic state although the succession of different variants is predictable from the GISAID website. The different transmissibility of dominant strain lead to no significant correlation between the new PANGO lineage and the number of new confirmed cases in the whole pandemic time. However, the pandemic trend of new confirmed cases was obviously opposite with the new PANGO lineage during a period with specific dominant strain (Figs. 3D and 4A). On the one hand, 20I (Alpha), and 21I or 21 J (both Delta) were the dominant variants of SARS-CoV-2 during December 2020 to April 2021, and April 2021 to November 2021, respectively; the regression line of new confirmed cases showed a rising trend at the same time. On the other hand, 21 L (BA.2) lost the dominant level gradually from April 2022, and the percentage of other variants (22 A-23B) was increasing; but the regression line of new confirmed cases went downwards meanwhile: both of them exhibited the negative correlation, which was further analyzed by Spearman rank correlation (P < 0.01) (Table 2), and indicate reference value for monitoring SARS-CoV-2 variants at the low epidemic state. The causes and implications of observed the negative correlation were explained as followed. The number of dominant viral variants decreased when the epidemic was at its peak, suggesting that the widespread virus strains had a strong dissemination advantage and formed a new epidemic wave to suppress the prevalence of new variants. On the other hand, the number of dominant virus variants increased when the epidemic experienced a trough, suggesting that many new variants were competing for dominance to spread widely and cause an epidemic wave. When the proportion of a certain variant increased and its transmission advantage was established, a new epidemic peak appeared. When the epidemic experienced a trough, the number of new dominant variants decreased simultaneously, which indicated the end of the epidemic. Currently, since the number of newly reported COVID-19 cases lacks reference importance, the number and change of dominant virus variants may be potential indicators for epidemic monitoring, early warning and prediction and even for evaluating the end of the pandemic. Homa Attar Cohen et al. also proposed that genomic surveillance of SARS-CoV-2 variants can provide essential, timely and evidence-based information for public health decision-making, including variant-associated environmental monitoring, laboratory assessment of variant evolution and phenotype and analyzing circulating variants with clinical data24.
Given that the data did not follow the normal distribution with K-S test, Kendall’s tau-b and Spearman rank correlation coefficient, the non-parametric statistical methods, were suitable for the correlation analysis of the data. Kendall’s tau-b rank correlation coefficient balances the data with many ties by losing some statistical information in ordered categorical data, while the Spearman rank correlation coefficient is not suitable for the data with many ties. However, there are no ties for new confirmed cases, and a few for new PANGO lineages. Therefore, the Spearman rank correlation coefficient is better for data analysis, which is easy and stable to perform, suitable for large samples, insensitive to outliers, and higher statistical information utilization.
Three intervals were divided for analysis based on variants transmissibility. The number of new cases has decreased a lot after 11/01/2023 and came to zero at 15/03/2023, and epidemic controls have been cancelled, which corresponded with 1st interval. The negative correlation between new PANGO lineages and new confirmed cases was not significant in the 1st interval due to its long-span time, which weakened the overall correlation. In contrast, this correlation was significant in the 2nd interval and 3rd interval, when the variant of Delta or Omicron was dominant, respectively. Besides, the COVID-19 pandemic has once entered a relatively stable low-prevalence state since the outbreak in the 2nd interval, while the 3rd interval included the complete process from the low-prevalence state to the peak of the Omicron epidemic, and then back to the low-prevalence state. The selection of three intervals met the requirements of this study with typical significance to analyze the low prevalence state of COVID-19 epidemic.
In addition, the number and density of dominant virus variants decreased substantially after the peak of the COVID-19 pandemic in July and August 2022. The dominant virus variants that prevailed in different regions were not identical because of multiple factors, such as the established immune barrier, and the prevention and control measures; rather, only a few variants, such as BQ.1, XBB.1, BA.5 and EG.5, accounted for the majority of dominant strains and were the most prevalent in terms of continuous or alternate transmission for more than 2 months when there were no emerging or competitive dominant variants. Regional differences in the spread and prevalence of SARS-CoV-2 led to the global distribution characteristics of viral variants and also determined the virus naming of the PANGO lineage and the Nextstrain clade, which were not synchronized but suitable for assess the status of COVID-19 epidemics in the low epidemic state. The prevalence of COVID-19 will continue to be low in the foreseeable future, and the pandemic will end without new dominant variants. It is recommended to encourage more nucleic acid testing for COVID-19 patients and further variants sequencing in the low prevalence state, which may help provide early warning of the epidemic situation. Surveillance of variants could be integrated into future pandemic preparedness frameworks and play an important role in different epidemics or prevalence periods.
Nevertheless, there are several limitations in monitoring and forecasting epidemic conditions through the use of dominant virus variants as indicators. The recorded data on dominant virus variants are derived from professional initiatives to detect and upload viral RNA sequences, which affects the effectiveness and sustainability of monitoring and early warning systems. Besides, more practice cases are needed to prove the effectiveness of the epidemic-predicting method by monitoring variants. Fortunately, monitoring virus variants effectively is progressing. For example, Antonella Pacini et al. established one method of droplet digital PCR to track and manage any new variant outbreak for global readiness against possible future SARS-CoV-2 waves25. Without effective monitoring and early warning tools in the low epidemic state, the variants monitoring method can provide valuable information for appropriate prevention and control measures to a certain extent.
In conclusion, monitoring reported SARS-CoV-2 variants may provide another reference for assessing the COVID-19 pandemic situation in the low epidemic state, regardless of the limited data on new cases, even though this method has some deficiencies and requires further optimization. However, the emerging epidemics, such as monkeypox, cholera and dengue fever, still lack effective methods for monitoring and risk assessment, which may result from their variability and immunogenicity.
Data availability
The datasets generated and/or analyzed during the study are available from the corresponding author on reasonable request.
References
Lazarus, J. V. et al. A multinational Delphi consensus to end the COVID-19 public health threat. Nature 611, 332–345. https://doi.org/10.1038/s41586-022-05398-2 (2022).
Mouliou, D. S. & Gourgoulianis, K. I. False-positive and false-negative COVID-19 cases: respiratory prevention and management strategies, vaccination, and further perspectives. Expert Rev. Respir. Med. 15, 993–1002. https://doi.org/10.1080/17476348.2021.1917389 (2021).
Organization, W. H. Statement on the fifteenth meeting of the IHR () Emergency Committee on the COVID-19 pandemic, (2005). https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic (2023).
Smith-Schoenwalder, C. WHO: The End to the COVID-19 Pandemic Is in Sight, < (2022). https://www.usnews.com/news/health-news/articles/2022-09-14/who-the-end-to-the-covid-19-pandemic-is-in-sight
Charumilind, S. et al. When will the COVID-19 pandemic end? Experts explain, (2021). https://www.weforum.org/agenda/2021/09/mckinsey-experts-offer-their-latest-analysis-on-when-covid-19-will-end/
Luan, J., Ba, J., Liu, B., Xu, X. & Shu, D. 2021–2022 monitoring, early warning, and forecasting of global infectious diseases. J. Biosaf. Biosecur. 4, 98–104. https://doi.org/10.1016/j.jobb.2022.06.001 (2022).
Ioannidis, J. P. A. The end of the COVID-19 pandemic. Eur. J. Clin. Invest. 52 (e13782). https://doi.org/10.1111/eci.13782 (2022).
Descary, M. H. & Froda, S. Estimating the basic reproduction number from noisy daily data. J. Theor. Biol. 549, 111210. https://doi.org/10.1016/j.jtbi.2022.111210 (2022).
Delamater, P. L., Street, E. J., Leslie, T. F., Yang, Y. T. & Jacobsen, K. H. Complexity of the basic reproduction number (R(0)). Emerg. Infect. Dis. 25, 1–4. https://doi.org/10.3201/eid2501.171901 (2019).
Roberts, M., Andreasen, V., Lloyd, A. & Pellis, L. Nine challenges for deterministic epidemic models. Epidemics 10, 49–53. https://doi.org/10.1016/j.epidem.2014.09.006 (2015).
Manabe, H. et al. Simple mathematical model for predicting COVID-19 outbreaks in Japan based on epidemic waves with a cyclical trend. BMC Infect. Dis. 24, 465. https://doi.org/10.1186/s12879-024-09354-5 (2024).
Tuli, S., Tuli, S., Tuli, R. & Gill, S. S. Predicting the growth and trend of COVID-19 pandemic using machine learning and cloud computing. Internet Things. 11, 100222. https://doi.org/10.1016/j.iot.2020.100222 (2020).
Li, B. et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat. Commun. 13, 460. https://doi.org/10.1038/s41467-022-28089-y (2022).
Guo, H. et al. SARS-CoV-2 Omicron: viral evolution, immune evasion, and alternative durable therapeutic strategies. Viruses 16 https://doi.org/10.3390/v16050697 (2024).
Committee, L. D. Rules for the Designation and Naming of Pango Lineages, < (2020). https://www.pango.network/the-pango-nomenclature-system/statement-of-nomenclature-rules/
O’Toole, Á., Pybus, O. G., Abram, M. E., Kelly, E. J. & Rambaut, A. Pango lineage designation and assignment using SARS-CoV-2 Spike gene nucleotide sequences. BMC Genom. 23, 121. https://doi.org/10.1186/s12864-022-08358-2 (2022).
Roemer., C., Hodcroft., E. B., Neher, R. A. & Bedford, T. SARS-CoV-2 clade naming strategy for 2022, (2022). https://nextstrain.org/blog/2022-04-29-SARS-CoV-2-clade-naming-2022
Hadfield, J. et al. Nextstrain: real-time tracking of pathogen evolution. Bioinf. (Oxford England). 34, 4121–4123. https://doi.org/10.1093/bioinformatics/bty407 (2018).
Flores-Vega, V. R. et al. SARS-CoV-2: evolution and emergence of new viral variants. Viruses 14 https://doi.org/10.3390/v14040653 (2022).
Liu, H., Wei, P., Kappler, J. W., Marrack, P. & Zhang, G. SARS-CoV-2 variants of concern and variants of interest receptor binding domain mutations and virus infectivity. Front. Immunol. 13, 825256. https://doi.org/10.3389/fimmu.2022.825256 (2022).
Shi, G. et al. Omicron Spike confers enhanced infectivity and interferon resistance to SARS-CoV-2 in human nasal tissue. Nat. Commun. 15, 889. https://doi.org/10.1038/s41467-024-45075-8 (2024).
Lee, J. A. et al. Development of a chimeric strain of Porcine reproductive and respiratory syndrome virus with an infectious clone and a Korean dominant field strain. J. Microbiol. 52, 345–349. https://doi.org/10.1007/s12275-014-4074-4 (2014).
Drake, J. W. Too many mutants with multiple mutations. Crit. Rev. Biochem. Mol. Biol. 42, 247–258. https://doi.org/10.1080/10409230701495631 (2007).
Cohen, A. Surveillance for variants of SARS-CoV-2 to inform risk assessments. Bull. World Health Organ. 101, 707–716. https://doi.org/10.2471/blt.23.290093 (2023).
Pacini, A. et al. Ready for new waves: optimizing SARS-CoV-2 variants monitoring in pooled samples with droplet digital PCR. Front. Public. Health. 11, 1340420. https://doi.org/10.3389/fpubh.2023.1340420 (2023).
Acknowledgements
The authors would like to thank their colleagues at Naval Medical University for their participation in this study.
Funding
This work was supported by the National Key Research and Development Program of China (2022YFC2602900).
Author information
Authors and Affiliations
Contributions
Bin Liu: literature search, figures, data interpretation, writing; Zhongliang Wang: data collection, data analysis, language editing; Shanshan Lu: data collection, data analysis, language editing; Zhongtian Qi: study design, data interpretation, writing; Zhijie Zhang: study design, data interpretation, writing; Jie Luan: study design, data interpretation, writing; Jianbo Ba: study design, data interpretation, writing. All the authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, B., Wang, Z., Lu, S. et al. Monitoring reported SARS-CoV-2 variants to assess the status of COVID-19 epidemics in the low epidemic state. Sci Rep 15, 10169 (2025). https://doi.org/10.1038/s41598-025-91308-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-91308-1